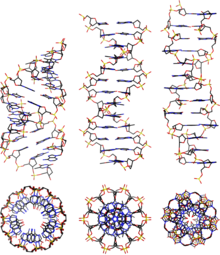
B-DNA
B-DNA is one of the possible double helix - structures of DNA .
properties
B-DNA is a right-handed double-stranded DNA double helix, which is more relaxed than the A-form and whose nucleobases are oriented orthogonally to the helix axis. B-DNA is the form of double-stranded DNA that is typically present under physiological conditions, e.g. B. in chromosomes in cells . Other DNA structures are e.g. B. A-DNA , Z-DNA , hairpin, triplex, cruciform, left-handed Z-form, tetraplex and A-motif . Due to the more relaxed shape, B-DNA has a smaller number of nucleobases per turn of the helix, a shallower large groove, a deeper small groove and is about 30% longer per turn than A-DNA.
Double-stranded DNA structures (dsDNA)
| geometry | A shape | B shape | Z shape |
|---|---|---|---|
| Helix direction of rotation | right-handed | right-handed | left-handed |
| Repetition unit | 1 bp | 1 bp | 2 bp |
| Rotation / bp | 33.6 ° | 34.6 ° | 60 ° / 2 |
| bp / turn | 11 | 10.4 | 12 |
| Inclination of the bp to the axis | + 19 ° | −1.2 ° | −9 ° |
| Length / bp along the axis | 2.4 Å (0.24 nm) | 3.4 Å (0.34 nm) | 3.7 Å (0.37 nm) |
| Length / turn | 24.6 Å (2.46 nm) | 33.2 Å (3.32 nm) | 45.6 Å (4.56 nm) |
| Bend (propeller twist) | + 18 ° | + 16 ° | 0 ° |
| Glycosyl angle | anti | anti | Pyrimidine: anti, purine: syn |
| Phosphate spacing | 5.9 Å | 7.0 Å | C: 5.7 Å, G: 6.1 Å |
| Glycosyl flexibility (sugar pucker) | C3'-endo | C2'-endo | C: C2'-endo, G: C3'-endo |
| diameter | 23 Å (2.3 nm) | 20 Å (2.0 nm) | 18 Å (1.8 nm) |
literature
- E. Girard, T. Prangé, AC Dhaussy, E. Migianu-Griffoni, M. Lecouvey, JC Chervin, M. Mezouar, R. Kahn, R. Fourme: Adaptation of the base-paired double-helix molecular architecture to extreme pressure . In: Nucleic acids research. Volume 35, number 14, 2007, pp. 4800-4808, doi: 10.1093 / nar / gkm511 , PMID 17617642 , PMC 1950552 (free full text).
- P. Cysewski: The post-SCF quantum chemistry characteristics of inter- and intra-strand stacking interactions in d (CpG) and d (GpC) steps found in B-DNA, A-DNA and Z-DNA crystals. In: Journal of molecular modeling. Volume 15, Number 6, June 2009, pp. 597-606, doi: 10.1007 / s00894-008-0378-9 , PMID 19039609 .
Web links
Individual evidence
- ↑ DL Beveridge, TE Cheatham, M. Mezei: The ABCs of molecular dynamics simulations on B-DNA, circa 2012. In: Journal of biosciences. Volume 37, Number 3, July 2012, pp. 379-397, PMID 22750978 , PMC 4029509 (free full text).
- ↑ AS Boyer, S. Grgurevic, C. Cazaux, JS Hoffmann: The human specialized DNA polymerases and non-B DNA: vital relationships to preserve genome integrity. In: Journal of molecular biology. Volume 425, number 23, November 2013, pp. 4767-4781, doi: 10.1016 / j.jmb.2013.09.022 , PMID 24095858 .
- ↑ J. Choi, T. Majima: Conformational changes of non-B DNA. In: Chemical Society reviews. Volume 40, Number 12, December 2011, pp. 5893-5909, doi: 10.1039 / c1cs15153c , PMID 21901191 .
- ↑ MC election, M. Sundaralingam: Crystal structures of A-DNA duplexes. In: Biopolymers. Volume 44, Number 1, 1997, pp. 45-63, PMID 9097733 , doi : 10.1002 / (SICI) 1097-0282 (1997) 44: 1 <45 :: AID-BIP4> 3.0.CO; 2- # .