Epigenetics
The epigenetics ( give a wiki. Ἐπί epi , to ', also' and genetics ) is the field of biology , which factors, which deals with the question of the activity of a gene and thus the development of the cell set temporarily. She investigates changes in gene function that are not based on changes in the sequence of deoxyribonucleic acid , for example through mutation or recombination, and are still passed on to daughter cells.
It is based on chemical changes in chromatin, the proteins that bind to DNA, or methylation of the DNA itself, which can influence sections or entire chromosomes in their activity. One also speaks of epigenetic change or epigenetic imprinting . Since the DNA sequence is not changed, epigenetic effects cannot be detected and observed in the genotype (DNA sequence), but in the phenotype . Typical examples of epigenetic processes are X-inactivation , genomic imprinting or the transcriptional memory of cells.
introduction

Genetic material is passed on during inheritance. Epigenetic fixation causes the totipotent cells to mature and specialize. The ripening process is usually irreversible. (Each arrow indicates a cell division. The cell is changed. These changes are passed on to the daughter cells with the genetic material. These are not changes in the sequence of the DNA.)
After fertilization, the egg cell divides . All daughter cells are equivalent up to the 8-cell stage. They are called totipotent , because each of them still alone is able to produce a complete organism. Then there are cells with a different internal program, whose development potential is limited from now on - i.e. H. more and more specialized - becomes. When the body is fully developed, most of the body cells are firmly programmed for their function (only the so-called adult stem cells retain a certain flexibility). The sequence of the genetic material remains unchanged (apart from a few random genetic changes = mutations ). The functional definition is carried out by various mechanisms, one of which is based on biochemical modifications at individual bases of the sequence or the histones packaging the DNA or both. Changes of this kind lead to certain areas of the genome being “shut down”, while others can be more easily transcribed ( rewritten in RNA for proteins ). These modifications look very different in body cells than in stem cells or in germ cells (egg cells and sperm ; cancer cells also usually have different [and thereby specific] modification patterns). The most important modifications are the methylation of cytidine bases on a CpG dinucleotide ( cytosine - guanosine - nucleotide - dimer , DNA methylation ) and the side chain methylation and acetylation of histones.
In addition to methylation, telomeres have an important epigenetic significance. Telomeres protect the ends of chromosomes from degradation during cell division. The enzyme telomerase ensures that the chromosomes remain intact. Psychological stress can reduce the activity of this enzyme, which can lead to an accelerated shortening of the telomeres in the aging process (Nobel Prize for Medicine 2009 to Elizabeth Blackburn ).
term
Epigenetic are all processes in a cell that are “additional” to the contents and processes of genetics. Conrad Hal Waddington first used the term epigenetics. In 1942 (when the structure of DNA was still unknown) he defined epigenetics as the branch of biology which studies the causal interactions between genes and their products which bring the phenotype into being (“the branch of biology that deals with the causal interactions between genes and their products that produce the phenotype ”). To distinguish it from the more general concept of gene regulation , today's definitions are usually more specific, for example: "The term epigenetics defines all meiotically and mitotically hereditary changes in gene expression that are not encoded in the DNA sequence itself." Other definitions, such as Adrian's Peter Bird , one of the pioneers of epigenetics, avoid the restriction to cross-generational transmission. Epigenetics describes “the structural adaptation of chromosomal regions in order to code, signal, or conserve altered states of activation.” In a review on epigenetics in bacteria , Casadesús and Low suggested using a preliminary definition, as long as it was not general accepted definition of epigenetics gives: "A preliminary definition could, however, be that epigenetics addresses the study of cell line formation by non-mutational mechanisms."
Epigenesis
The term epigenesis describes the gradual processes of embryonic morphogenesis of organs . These are based on mechanisms at the level of cells and cell associations, these are Turing mechanisms or general pattern formation processes in biology. Examples of this can be found in the explanation of the embryonic extremity development in vertebrates.
Associated terms
The epigenetic processes include paramutation , bookmarking , imprinting , gene silencing , X-inactivation , the position effect , reprogramming , transvection , maternal effects (paternal effects are rare because there is much less non-genetic material involved The sperm is “inherited”), the process of carcinogenesis , many effects of teratogenic substances, the regulation of histone modifications and heterochromatin as well as technical limitations in cloning .
Epigenetics versus Genetics
The term epigenetics can be understood by considering the process of inheritance:
- Before cells divide , the genetic material is doubled. Half of the doubled genome is then transferred to one of the two daughter cells. In the sexual reproduction of humans, the reproductive , half of the maternal genome and by the sperm half are combined the paternal genome together from the egg.
- Molecular genetics describes the genetic material as a double helix made up of two deoxyribonucleic acid strands, the backbone of which consists of a phosphate-deoxyribose sugar polymer each. The genetic information is determined by the order of the four bases adenine (A), cytosine (C), guanine (G) and thymine (T), which are each attached to one of the deoxyribose sugars.
- The bases of one strand almost always pair with a matching base of the second strand. Adenine pairs with thymine, and cytosine pairs with guanine.
- The genetic information is anchored in the order of the building blocks A, C, G, T (the base sequence).
Some inheritance phenomena cannot be explained with the DNA model just described:
- During cell differentiation , daughter cells with a different function arise in the course of cell division, although the genetic material is the same in all cells. Establishing the functional identity of a cell is a subject of epigenetics.
- There are properties that are only “inherited” from the father (paternal), just as there are properties that only come from the mother (maternal) and are not related to the base sequence. Disturbances in this condition lead to serious illnesses.
- When converting functionally established cells (terminally differentiated cells) back into undifferentiated cells, which can develop again into different cells and which are used in the cloning of individuals (e.g. from Dolly ), epigenetic fixations must be broken so that a Cell does not remain restricted to a single function, but can acquire and inherit all or many functions again.

The DNA is wrapped around the core of eight histone subunits (two each H2a, H2b, H3 and H4) and makes about 1.7 turns. Histone 1 (H1) binds to the piece of DNA between two nucleosomes. The ends of the histones are available for epigenetic modification: methylation, acetylation or phosphorylation. This affects the compression or expansion of the chromatin.
Histones and their role in epigenetic fixation
DNA is not naked in the cell nucleus, but is bound to histones . Eight different histone proteins, two molecules each of histone 2A , histone 2B , histone 3 and histone 4 form the core of a nucleosome, onto which 146 base pairs of a DNA strand are coiled. The ends of the histone strands protrude from the nucleosome and are the target of histone-modifying enzymes. The known modifications are mainly methylations and acetylations on lysine, histidine or arginine, and phosphorylations on serines. It also plays a role whether the lysine side chain has one, two or three methyl groups. Comparative analysis postulates a type of “ histone code ” that is supposed to be directly related to the activity of the gene bound by the histones.
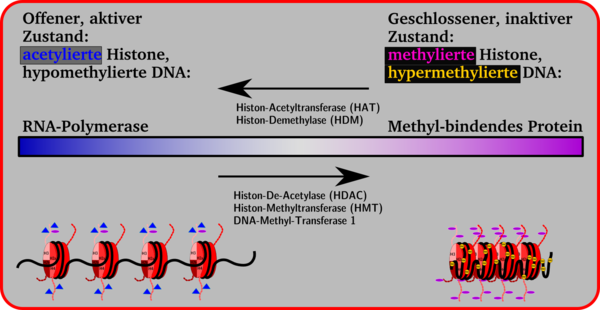
The histone side chains in the nucleosomes can be changed enzymatically. This changes the volume of a gene segment. Smaller volumes, closed conformation, chromosome condensation and inactivity of a gene are on the one hand, larger volumes, open conformation and gene activity on the other. A transition is possible between the two sides, which is brought about by the attachment and cleavage of methyl groups on cytidine bases, by methylation, demethylation, acetylation or deacetylation with the aid of enzymes.
In general, it can be said that the attachment of acetyl groups to the lysine side chains of the histones leads to the opening of the nucleosome conformation, making the gene available for transcription by the RNA polymerase. By an increased attachment of methyl groups to lysine side chains, proteins are attached such. B. the methyl-binding protein MeCB , which suppresses gene expression . These repressor proteins cause the histone conformation to close and transcription is no longer possible.
Methods of Epigenetic Research
Restriction endonucleases that only cut on demethylated CG dimers
HpaII (the second restriction endonuclease from Haemophilus parainfluenza ) only cuts CCGG palindromes if the CG dimers are not methylated, compared to BsiSI (from Bacillus), which also cuts methylated C me CGG palindromes. Tryndiak and co-workers show that in cells on the way to breast cancer, a progressive global loss of DNA methylation is associated with a misdirected formation of DNMT1, me CG-binding proteins and changes in the histones.
Bisulfite sequencing
By treating DNA with sodium hydrogen sulfite (old name "bisulfite"), cytosine (C) is converted into uracil (U). In a subsequent sequencing , you will find a U / T at the places where there was a C before. Since bisulfite-treated DNA is very labile, the gene that you want to analyze is amplified again using PCR . In the subsequent sequencing, T or TG (thymine-guanosine dimers) are identified where cytosine or CG dimers existed in the untreated DNA.
For epigenetic analysis it is important that only non-methylated C bases are converted, while me C in CG dimers are not converted to thymine. This method can therefore be used to precisely analyze which CG dimers were methylated in a particular cell. By cloning the bisulfite-treated gene piece that you want to analyze after the PCR amplification and sequencing different clones, you can get an estimate of whether a certain CG dimer was not, completely or only partially methylated. In the pyrosequencing method, this process is refined and allows more precise quantitative statements: for example, the degree of severity of tumor degeneration can be compared with the degree of methylation of CG islands of individual so-called tumor suppressor proteins and it is established that in certain tumors of the blood-forming system (hematopoietic System) certain me CG dimers are more and more methylated with increasing severity.
Chromatin Immunoprecipitation
With this method you can determine whether a certain protein binds to a given piece of DNA: By treating the cells or biological tissue with formaldehyde , the binding proteins are covalently linked to the DNA. The DNA extracted from the cells is then fragmented into fragments of 50 to 1000 base pairs by treatment with ultrasound, on which the bound proteins remain. In a next step, the protein of interest is extracted with an antibody together with the DNA fragments bound to it, and the covalent bond between protein and DNA is then dissolved again by heat treatment in 300 mM saline solution. The DNA fragments thus separated can then be identified or quantified (using gene-specific PCR or genome-wide NGS ). From the frequency with which a certain DNA fragment is identified, it can be concluded whether or how strongly the protein in the living cell was associated with the relevant DNA segment. Depending on which protein you are trying to precipitate with antibodies, you can z. B. say:
- The RNA polymerase bound to the gene, so it was transcribed, the gene was active.
- The me CG-binding protein (MeCP) was linked to the gene, so this was not transcribed, and was sedated (Engl. Silencing ).
Electrophoretic Mobility Shift Assay
The different DNA molecules show different behavior in gel electrophoresis .
Epigenomics and methods of epigenetic research
Epigenomics or epigenomics is a branch of epigenetics that aims to investigate the most complete set of epigenetic modifications possible on the genetic material of a cell. Such related sets of epigenetic modifications are called epigenomes .
The term epigenomics was formed in analogy to other -omics , such as genomics and proteomics , and became popular when methods became available to study epigenetic modifications on a larger scale. The initiation of the Human Epigenome Project in 1999 made a significant contribution to this.
Epigenomics and epigenetics are not mutually exclusive. Comprehensive and efficient research into epigenetics on a global level is made possible by high-throughput methods. The use of the term epigenome research or epigenomics characterizes this approach more specifically than the use of the term epigenetics.
Two of the most characterized epigenetic markers are DNA methylation and histone modifications. An overall determination of DNA methylation at a particular time, in a particular tissue, etc. is often referred to as a methylome or DNA methylation pattern, and a related set of histone modifications is often called a histone code . Both methylomes (or DNA methylation patterns) and histone codes are partial epigenomes.
Bisulfite sequencing and ChIP-Seq are examples of the investigation of epigenomes . Bisulfite sequencing enables a comprehensive analysis of "methylomes" (DNA methylation patterns) and ChIP-Seq can be used for the interaction of histones with DNA.
It should be noted that although epigenome research can be viewed as a sub-area of epigenetics, it is a very interdisciplinary subject that has, for example, intersections with genetics , molecular biology, all omics areas, systems biology and bioinformatics .
Epigenetic changes in the life course
Epigenetics is not limited to cases of inheritance. Epigenetic research results in connection with persistent changes in the life course as well as in connection with the development of diseases are receiving increasing attention. In 80 identical twins, for example, it was possible to demonstrate that they still correspond epigenetically to a high degree at the age of three, but no longer at the age of 50 if they have spent little life together and / or have had different medical and health histories . The degree of methylation was up to two and a half times higher in a twin, both in absolute numbers and in terms of the distribution of the epigenetic markers. Older twins, despite their genetic identity, are epigenetically more different the more different the life of the twins is. The reason lies not only in the experienced environment but also in the inaccuracy in the transmission of methyl group patterns with every cell division. Creeping changes therefore add up more and more over the course of a lifetime.
The change in the diet of worker bees after the first few weeks of the larval stage to a simple pollen and honey diet compared to the queen causes a high-grade epigenetic reprogramming of the larval genome. More than 500 genes have been identified that are affected by the environmentally specific methylation changes. Not only activation or non-activation of genes is the result of the change in diet, but also alternative splicing and modified gene products .
Epigenetic changes as an explanation of diseases
The explanation of stress factors is a focus of epigenetic research. Individuals with early traumatic life experiences, for example caused by a lack of maternity care from rat mothers, were used for this. Stress sets in motion a cascade of hormone releases to control it, the chain of which begins in the hypothalamus , part of the diencephalon. It could be shown that a glucocorticoid gene exhibits strikingly different methylations in the individuals concerned. Accordingly, the gene is inhibited in the presence of a stressful past. The gene product in the adrenal cortex as the end station of the hormone chain is different as a result. More than 900 genes are up or down regulated in the brain as a result of maternal behavior.
The results could also be confirmed in humans. In humans, the receptor gene in the hippocampus largely corresponds to that of other mammals. Epigenetic changes are therefore similar to those in rats. A study of suicide candidates divided those affected into two groups, those with experience of abuse in childhood and those without. Only in the candidates with a history of abuse was the receptor gene blocked with methylation. According to the same epigenetic pattern, trauma that the mother experiences during pregnancy can even have lasting consequences for the child-to-be, which last for decades for the child. In a study from the Netherlands it was shown that children of mothers who suffered from famine very early in their pregnancy showed a significantly increased risk of schizophrenia and cardiovascular disease in the course of their life and at the same time showed changes in the methylation pattern of the Igf2 gene carried.
In mice, regular cocaine use changes the pattern of epigenetic markers of a few hundred genes in the reward center of the brain. This increases the sensitivity to the effects of the drug and increases the risk of addiction.
The magnitude of epigenetic changes in the course of life is many times higher than that of genetic mutations. Science therefore expects further new answers in the future to a multitude of diseases in the aging organism that cannot be explained genetically today, including schizophrenia , Alzheimer's disease , cancer, old-age diabetes , nervous diseases and others.
“Inheritance” of epigenetic influences?
From the findings on epigenetic changes, especially in popular science, parallels to Lamarckism are repeatedly drawn and a contradiction to classical genetics is seen. So far, however, there is very little evidence that learned and acquired skills can be passed on from one generation to another via the germ cells. Passing on to the next generation is not yet proof of genetic manifestation, and the term “generation” is often misinterpreted as the beginning of an individual cycle.
An inheritance of epigenetic imprints was suggested in 2003 by Randy Jirtle and Robert Waterland using mouse experiments. Female agouti mice were given a specific set of nutrients before mating and during pregnancy. It turned out that the majority of the offspring did not show the typical phenotype .
In a human study, the two geneticists Marcus Pembrey and Lars Olov Bygren and colleagues examined various factors that provided information about food availability and deaths in the small Swedish town of Överkalix . It found that most people whose grandparents had enough to eat during childhood developed diabetes as they grew older . However, the disease occurred according to a certain pattern, which suggests epigenetic changes on the sex chromosomes . For example, in families in which the grandfather had fed well or excessively, only the male grandchildren were affected of all grandchildren.
According to a hypothesis by William R. Rice, Urban Friberg and Sergey Gavrilets from 2012, the development of human homosexuality could be caused by epigenetic "inheritance". Thus, in some individuals, the mother's sexual preference would be passed on to the son and the father's preference to the daughter. This happens when the epigenetic DNA markings ("Epi-Marks") in genes that are responsible for sexual orientation are retained in the germ cells. If, for example, these epigenetic DNA markings in the unfertilized egg cell (i.e. the maternal germ cell) are not completely reset, an embryo could indeed develop male reproductive organs (if it has inherited the XY genotype), the sexual orientation would be towards the male gender but similar to the mother's. According to this hypothesis, human homosexuality is innate. The hypothesis explains why the incidence of homosexuality in humans remains statistically stable over time. However, the authors of Rice also write that it is only a hypothesis, but so far there is no empirical evidence for a connection between homosexuality and epigenetics. A critical analysis of the Rice et al. made by Heinz J. Voss .
Epigenetics in bacteria
While epigenetics is of fundamental importance in eukaryotes , especially in multicellular animals ( Metazoa ), it plays a different role in bacteria. Most importantly, eukaryotes use epigenetic mechanisms primarily to regulate gene expression , which bacteria rarely do. However, the use of post-replicative DNA methylation for the epigenetic control of DNA-protein interactions in bacteria is widespread. In addition, bacteria use DNA adenine methylation (instead of DNA cytosine methylation) as an epigenetic signal. DNA adenine methylation is important in bacterial virulence in organisms such as Escherichia coli , Salmonella , Vibrio , Yersinia , Haemophilus, and Brucella . In the alphaproteobacteria , the methylation of adenine regulates the cell cycle and connects gene transcription with DNA replication. In gammaproteobacteria , adenine methylation provides signals for DNA replication, chromosome segregation, mismatch repair, packaging of bacteriophages , transposase activity and regulation of gene expression. There is a genetic switch that controls Streptococcus pneumoniae ( pneumococci ) and allows the bacterium to randomly change its properties so that one of six alternative states occurs. That could pave the way for improved vaccines. Each shape is generated randomly by a phase-variable methylation system. The ability of the pneumococci to cause fatal infections is different in each of these six conditions. Similar systems exist in other types of bacteria.
literature
Monographs
- Jan Baedke: Above the Gene, Beyond Biology: Toward a Philosophy of Epigenetics. University of Pittsburgh Press, Pittsburgh 2018.
- Joachim Bauer : The body's memory: how relationships and lifestyles control our genes. Eichborn, Frankfurt am Main 2002; Expanded paperback edition: Piper, Munich 2004 (10th edition 2007), ISBN 978-3-492-24179-3 .
- Bernhard Kegel : Epigenetics. How experiences are inherited . Dumont, Cologne 2009, ISBN 978-3-8321-9528-1 .
- Oscar Hertwig : Biological problem of today: preformation or epigenesis? The basis of a theory of organic development. Heinemann, London 1896.
- Peter Spork : The second code. Epigenetics - or how we can control our genetic makeup. Rowohlt, Reinbek 2009, ISBN 978-3-498-06407-5 .
- Wolfgang Wieser : Brain and Genome: A New Screenplay for Evolution. Beck, Munich 2007, ISBN 3-406-55634-5 .
Essays
- Bradbury, J. (2003): Human Epigenome Project-Up and Running. In: PLoS Biol . Vol. 1, p. E82 doi: 10.1371 / journal.pbio.0000082 ; PDF (free full text access).
- Costa, FF. (2008): Non-coding RNAs, epigenetics and complexity . In: Gene Vol. 410, No. 1, pp. 9-17, PMID 18226475 , doi: 10.1016 / j.gene.2007.12.008 .
- Jablonka, E. and Lamb, MJ. (2002): The Changing Concept of Epigenetics . In: Annals of the New York Academy of Sciences Vol. 981, pp. 82-96, PMID 12547675 .
- Delcuve, GP. et al. (2009): Epigenetic control . In: J Cell Physiol . Vol. 219, No. 2, pp. 243-250, PMID 19127539 .
- Marmorstein, R. and Trievel, RC. (2009): Histone modifying enzymes: structures, mechanisms, and specificities . In: Biochimica et Biophysica Acta (BBA) Vol. 1789, No. 1, pp. 58-68, PMID 1872256 , doi: 10.1016 / j.bbagrm.2008.07.009 .
- Morgan, HD. et al. (2005): Epigenetic reprogramming in mammals . In: Human molecular genetics . (Hum Mol Genet.) Vol. 14, No. 1, R47-58, PMID 15809273 ; PDF (free full text access)
- Peter Spork et al. (2016): 'Inherited Fear'; 'Traces in the family tree'; 'Those who mourn can let go of suffering'; picture of science 2/2016, pp. 16-25.
- Jan Baedke (2019): Philosophical Problems of Epigenetics. Report. In: Information Philosophie Heft 2/2019, pp. 22–31. Bibliography in Information Philosophy.
Press
- Breaking the Evil Spell . In: Der Spiegel . No. 32 , 2008 ( online ).
Web links
Portals
- The Epigenome Network of Excellence (contains references to recent publications )
- Epigenetics: Portal of Epigenetic Research
- Public European Epigenome Network (multilingual)
- Epigenetics newsletter website
Individual contributions
- Epigenetics: how the environment and behavior control genes. Audio on: funkkolleg-biologie.de , November 18, 2017
- Traces of ancestors on our genetic makeup (NZZ 2012)
- “Epigenetics” from Planet Wissen , accessed on May 26, 2015.
- Why hasn't all the brilliant geneticists noticed in a hundred years? published in Spektrum March 26, 2014
- Stephanie Lahrtz: Gene packaging influences behavior. Reprogrammed ants. Neue Zürcher Zeitung, January 2, 2016, accessed on January 2, 2016 .
Individual evidence
- ↑ Rudolf Hagemann: Epigenetics and Lamarckism have nothing in common! In: Laborjournal 4/2009; P. 12
- ^ Benjamin Lewin: Genes. Textbook of molecular genetics , 2nd edition, VCH, Weinheim 1991, p. 885
- ↑ Agustina D'Urso, Jason H. Brickner: Epigenetic transcriptional memory . In: Current Genetics . tape 63 , no. 3 , November 2, 2017, ISSN 0172-8083 , p. 435-439 , doi : 10.1007 / s00294-016-0661-8 , PMID 27807647 , PMC 5413435 (free full text) - ( springer.com [accessed July 30, 2019]).
- ↑ "The term epigenetics defines all meiotically and mitotically heritable changes in gene expression that are not coded in the DNA sequence itself." In: Gerda Egger et al .: Epigenetics in human disease and prospects for epigenetic therapy . Nature 429, pp. 457-463 (2004)
- ^ "... the structural adaptation of chromosomal regions so as to register, signal or perpetuate altered activity states." In: Adrian Peter Bird : Perceptions of epigenetics. In: Nature. Volume 447, number 7143, May 2007, pp. 396-398, doi: 10.1038 / nature05913 , PMID 17522671 .
- ↑ "However, a tentative definition may be that epigenetics addresses the study of cell lineage formation by non-mutational mechanisms." In: Josep Casadesús and David A. Low: Programmed heterogeneity: epigenetic mechanisms in bacteria. In: The Journal of biological chemistry. Volume 288, number 20, May 2013, pp. 13929-13935, doi : 10.1074 / jbc.R113.472274 , PMID 23592777 , PMC 3656251 (free full text) (review).
- ↑ Figure by Clapier et al., Protein database 2PYO ; Clapier, CR. et al. (2007): Structure of the Drosophila nucleosome core particle highlights evolutionary constraints on the H2A-H2B histone dimer . In: Proteins 71 (1); 1-7; PMID 17957772 ; PMC 2443955 (free full text)
- ↑ Tryndiak, VP. et al. (2006): Loss of DNA methylation and histone H4 lysine 20 trimethylation in human breast cancer cells is associated with aberrant expression of DNA methyltransferase 1, Suv4-20H2 histone methyltransferase and methyl-binding proteins . In: Cancer Biol Ther . 5 (1), 65-70; PMID 16322686 ; PDF (free full text access)
- ↑ "... The Human Epigenome Project, for example, was established in 1999, when researchers in Europe teamed up to identify, catalog and interpret genomewide DNA methylation patterns in human genes. ..." In: L. Bonetta: Epigenomics: Detailed analysis. In: Nature. Volume 454, 2008, p. 796, doi : 10.1038 / 454795a .
- ↑ "... Over time, the field of epigenetics gave rise to that of epigenomics, which is the study of epigenetic modifications across an individual's entire genome. Epigenomics has only become possible in recent years because of the advent of various sequencing tools and technologies , such as DNA microarrays, cheap whole-genome resequencing, and databases for studying entire genomes (Bonetta, 2008) ... " In: L. Bonetta:" Epigenomics: The new tool in studying complex diseases. " Nature Education. Volume 1, 2008, p. 178, web link .
- ^ KA Janssen, S. Sidoli, BA Garcia: Recent Achievements in Characterizing the Histone Code and Approaches to Integrating Epigenomics and Systems Biology. In: Methods in enzymology. Volume 586, 2017, pp. 359-378, doi : 10.1016 / bs.mie.2016.10.021 , PMID 28137571 , PMC 5512434 (free full text) (review).
- ↑ Mario F. Fraga, Esteban Ballestar, Maria F. Paz, Santiago Ropero, Fernando Setien, Maria L. Ballestar, Damia Heine-Suñer, Juan C. Cigudosa, Miguel Urioste, Javier Benitez, Manuel Boix-Chornet, Abel Sanchez-Aguilera , Charlotte Ling, Emma Carlsson, Pernille Poulsen, Allan Vaag, Zarko Stephan, Tim D. Spector, Yue-Zhong Wu, Christoph Plass, and Manel Esteller. Epigenetic differences arise during the lifetime of monozygotic twins. Proceedings of the National Academy of Sciences . 2005. July 26, 2005. Vol. 102. No. 30th
- ↑ Frank Lyko, Sylvain Foret a. a .: The Honey Bee Epigenomes: Differential Methylation of Brain DNA in Queens and Workers. In: PLoS Biology. 8, 2010, p. E1000506, doi: 10.1371 / journal.pbio.1000506 .
- ^ Carlson NR: Physiology of Behavior , 11th. Edition, Allyn & Bacon, New York 2010, ISBN 978-0-205-23939-9 , p. 605.
- ↑ Belanoff JK, Gross K, Yager A, Schatzberg AF: Corticosteroids and cognition . In: Journal of Psychiatric Research . 35, No. 3, 2001, pp. 127-45. doi : 10.1016 / S0022-3956 (01) 00018-8 . PMID 11461709 .
- ↑ Sapolsky RM: Glucocorticoids, stress and exacerbation of excitotoxic neuron death . In: Seminars in Neuroscience . 6, No. 5, October 1994, pp. 323-331. doi : 10.1006 / smns.1994.1041 .
- ↑ Patrick O McGowan, Aya Sasaki et al. a .: Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. In: Nature Neuroscience. 12, 2009, p. 342, doi: 10.1038 / nn.2270 .
- ↑ Violence against pregnant women changes the genetics of children. In: Gesundheitsindustrie-bw.de. BIOPRO Baden-Württemberg GmbH, July 25, 2011, accessed on February 16, 2017 .
- ↑ E. Susser, R. Neugebauer, HW Hoek, AS Brown, S. Lin, D. Labovitz, JM Gorman: Schizophrenia after prenatal famine. Further evidence. In: Archives of general psychiatry. Volume 53, Number 1, January 1996, pp. 25-31, PMID 8540774 .
- ↑ RC Painter, SR de Rooij, PM Bossuyt, TA Simmers, C. Osmond, DJ Barker, OP Bleker, TJ Roseboom: Early onset of coronary artery disease after prenatal exposure to the Dutch famine. In: The American Journal of Clinical Nutrition. Volume 84, Number 2, August 2006, pp. 322-327, doi : 10.1093 / ajcn / 84.1.322 , PMID 16895878 .
- ↑ BT Heijmans, EW Tobi, AD Stein, H. Putter, GJ Blauw, ES Susser, PE Slagboom, LH Lumey: Persistent epigenetic differences associated with prenatal exposure to famine in humans. In: Proceedings of the National Academy of Sciences . Volume 105, number 44, November 2008, pp. 17046-17049, doi : 10.1073 / pnas.0806560105 , PMID 18955703 , PMC 2579375 (free full text).
- ↑ Maze, L. Nestler, EJ: The Epigenetic Landscape of Addiction . In: Annals of the New York Academy of Sciences 1216, pp. 99-113, 2011.
- ↑ Tsankova, N. et al. Epigenetic Regulation in Psychiatric Disorders. in. Nature Reviews Neuroscience 8, pp. 355-367. 2007
- ↑ spiegel.de: Epigenetics: Mothers Can Pass On Experiences , February 4, 2009.
- ^ Nutrition up-to-date, (The Austrian Nutrition Society publishes the magazine “Nutrition up-to-date” four times a year.) 1/2007 pp. 1-6
- ^ Waterland, RA. and Jirtle, RL. (2003): Transposable elements: targets for early nutritional effects on epigenetic gene regulation . In: Mol Cell Biol . 23 (15); 5293-5300; PMID 12861015 ; PMC 165709 (free full text)
- ↑ ME Pembrey, LO Bygren, G. Kaati, p Edvinsson, K. North Stone, M. Sjöström, J. Golding: Sex-specific, male-line transgenerational responses in humans. In: European journal of human genetics: EJHG. Volume 14, Number 2, February 2006, pp. 159-166, doi: 10.1038 / sj.ejhg.5201538 , PMID 16391557 ; PDF (free full text access)
- ↑ scinexx.de: Environment or genes? The twins ' answer , February 26, 2013.
- ^ William R. Rice, Urban Friberg, and Sergey Gavrilets: Homosexuality as a Consequence of Epigenetically Canalized Sexual Development . The Quarterly Review of Biology, Vol. 87, No. 4, December 2012, PMID 23397798 ; ... The authors used "published data ... to develop a new model for homosexuality."
- ↑ a b Heinz J. Voss : Epigenetics and Homosexuality (PDF; 564 kB).
- ^ Jörg Tost: Epigenetics . Caister Academic Press, Norfolk, England 2008, ISBN 1-904455-23-9 .
- ↑ Casadesús J, Low D: Epigenetic gene regulation in the bacterial world . In: Microbiol. Mol. Biol. Rev. . 70, No. 3, September 2006, pp. 830-56. doi : 10.1128 / MMBR.00016-06 . PMID 16959970 . PMC 1594586 (free full text).
- ↑ Manso AS, Chai MH, Atack JM, Furi L, De Ste Croix M, Haigh R, Trappetti C, Ogunniyi AD, Shewell LK, Boitano M, Clark TA, Korlach J, Blades M, Mirkes E, Gorban AN, Paton JC , Jennings MP, Oggioni MR: A random six-phase switch regulates pneumococcal virulence via global epigenetic changes . In: Nature Communications . 5, September 2014, p. 5055. doi : 10.1038 / ncomms6055 . PMID 25268848 . PMC 4190663 (free full text).