Mitochondrial Eve

The mitochondrial Eve is a term from the archaeogenetics and referred to a woman, from whose mitochondrial DNA (mtDNA) mitochondrial DNA of all living humans by direct lineage emerged. Its male counterpart is the Adam on the Y chromosome .
theory
If you look at different genes (or other delimitable sections of the genome) in different individuals, you will usually find that there are slightly different variants in different individuals, which are called alleles ( polymorphism of the genome). If the individuals are concerned with corresponding, briefly homologous genes or DNA segments, these variants can only have arisen because an original sequence has gradually changed through mutations - this already corresponds to the definition of homologous genes. Each (homologous) DNA segment can be traced back to an original sequence from which today's diversity has gradually developed. If this process is followed over time, a pattern of splitting processes emerges, each of which is based on a mutation. Assuming, instead, the present sequences and tries to reconstruct their formation, each of these splits corresponds in retrospect a "coalescence" (English coalescence) of the respective sequences. Corresponding analyzes are therefore referred to as coalescence analyzes.
In humans, every chromosome, and therefore every gene, is in principle available in two copies, one from the mother and one from the father ( diploidy ). Exceptions to this are, in addition to the sex chromosomes , the independent genes of the mitochondria - these are organelles that primarily serve as energy suppliers (“power plants”) for the cell. Due to the special structure of the sperm , all mitochondria, both in men and women, come from the egg cell and thus carry the maternal genome. The mitochondrial genes of every individual thus all have the same haplotype . By means of coalescence analysis of the mitochondrial DNA , a DNA sequence can be determined mathematically, to which today's range of variation can be traced back. The wearer of this particular sequence is called, somewhat boldly, "mitochondrial Eva".
The existence of this ancestor is trivial and does not involve any special knowledge - it arises automatically from the fact that all people, indeed all living beings, are ultimately related to one another and therefore must have had a common ancestor at some point. Logically, the "mitochondrial Eve" would not even have to have belonged to our species, because speciation goes back to a population that could already have possessed a considerable polymorphism, so that the common ancestor from whom the mitochondrial sequence originates is already one Predecessor species would originate. Since the polymorphism and mutation rate are different in different genes, but also simply by chance, a different common ancestor emerges when looking at other genes, which can be much younger or older than the "mitochondrial Eve". It only becomes scientifically interesting if further knowledge can be added to this trivial basic statement, for example on the place or time of the split.
Under ideal conditions (infinite population size, unlimited mix, adaptively equivalent alleles, no mutations), every allele present in the population would remain permanently unchanged at a completely unchanged frequency ( Hardy-Weinberg equilibrium ). Of course, this is never the case in real populations. With a limited population size, alleles will simply become more or less common by chance, due to different numbers of offspring of individual individuals; this phenomenon is known as genetic drift . As a result, the lifespan of alleles in real populations is limited by genetic drift, even if they are completely equivalent to one another (neutral). It is easy to see that the genetic drift must be stronger the smaller the population size. The random path (mathematically a Markov chain ) leads, regardless of the initial size, every allele present in a population to extinction sooner or later - in the absence of further mutations until only one is left (called “fixation” of the allele). In the case of haploid inheritance (as with the mitochondrion), the probability of coalescence in the previous generation is 1 divided by the population size N (actually the "effective" population size , which is influenced by different pairing probabilities in addition to the number of individuals). This results in an expected value for the coalescence time of twice the population size. With a population size of 100 individuals, a common ancestor would be expected around 200 generations ago. In real populations, however, the influence of mutations that create new alleles must be taken into account.
The mitochondrial Eve was neither the first woman nor the only woman at some point of the past. Eva had many contemporaries; however, the mitochondrial lineages of the other women died out, while Eva's survived (in most of her offspring, however, with more or less mutations). The time and place of life of this ancestor can be narrowed down very precisely with the help of the analysis of the mtDNA of a representative number of individuals living today.
meaning
A number of properties make mtDNA a valuable tool for researching human ancestry:
- In comparison to the DNA of the cell nucleus , the mtDNA shows a higher and more constant mutation rate .
- Since mtDNA is only passed on from the mother, the effective population size is only ¼ as large as that of autosomal DNA, in which both parents have two copies each. Correspondingly, the fixation of alleles is also about four times as fast. The alleles of human mtDNA are therefore much younger than those of autosomal DNA and are very suitable for research into young human history, such as the colonization of Eurasia.
- The mutation rate and gene drift of mtDNA, which is higher than that of autosomal DNA, mean that the frequency of the alleles fluctuates much more from one subpopulation to another. From these differences, statements about ancestry, migration, displacement or mixing of populations can be derived much more easily than with the geographically more homogeneous autosomal DNA.
- Since a cell contains many mitochondria and there are several copies of the mtDNA in each, enough mtDNA can often be extracted from fossils (e.g. Neanderthal bone tissue) for analysis, while the DNA of the cell nucleus is far more rarely sufficiently complete.
- The lack of recombination in the inheritance of mtDNA enables statements to be made about specific properties of the female lineage.
Where and when did Eva live?
The first studies of the variation in human mitochondrial DNA were carried out as early as 1983. The theory became known through a publication by Rebecca L. Cann , Allan Wilson and Mark Stoneking (1987).
Cann et al. (1987) extracted mtDNA from the placenta of women from different parts of the world. Instead of sequencing the mtDNA, they conducted a restriction fragment length polymorphism (RFLP) study. They arranged the mtDNAs according to their similarity on a family tree and finally determined the root of the family tree. Two main branches branched off from the determined root: on one there were only Africans, on the other people from all parts of the world. From this the authors concluded that mitochondrial Eve must have lived in Africa.
They also tried to use a molecular clock to determine when mitochondrial Eve was alive. In 1987, data on the mitochondrial DNA (mtDNA) of fish, birds and some mammal species were already available. These data showed that mtDNA changes approximately 2-4% per million years. These data were used to calibrate the molecular clock. Since the human mitochondrial DNAs differed by only 0.57% on average in the study, it was concluded that the mitochondrial Eve must have lived only about 200,000 years ago.
Since the fixation of a lineage in an expanding population is unlikely - so the authors' argument - mitochondrial Eve must have lived before the ancestors of all humans living today left Africa. For the authors this is a clear indication of the Out-of-Africa theory . The hypothesis of the “ multiregional origin of modern man ” was rejected by the authors, as the mitochondrial Eve would have had to be much older for this ( Homo erectus had left Africa almost 2 million years ago).
The publication was heavily criticized from the start. Many points of criticism appeared to be justified:
- Of the 147 people in the study, only 2 of the 20 “Africans” were really from sub-Saharan Africa; the other 18 were African American .
- The method of generating the family tree did not necessarily yield the statistically most favorable tree.
- To find the root of the tree, the root was placed in the middle of the longest branch ( midpoint rooting ). This can lead to an incorrect position of the root, e.g. B. when the rate of evolution in Africa is higher.
- RFLP is not very useful for determining mutation rates, which is important for a molecular clock .
- Bad statistical analysis.
So the (serious) criticism was not directed against the concept of mitochondrial Eve in general . It was clear to everyone who was familiar with the subject that this woman must have lived somewhere and at some point. Only the scientific approach of the authors was criticized.
However, later, improved studies confirmed and supported the most important statements of Cann et al. (1987). For example, Ingman et al. (2000) a new, improved study by:
- They took samples from 53 people, 32 of them from different parts of sub-Saharan Africa.
- They sequenced the entire mtDNAs, but excluded the rapidly evolving D-loop region for analysis .
- The root of the family tree was determined using the mtDNA of a chimpanzee ( outgroup rooting ).
The results of this improved study were even clearer than the 1987 study:
- Complete separation of Africans and non-Africans.
- The first three branches of the family tree led only to Africans, the fourth led to Africans and non-Africans.
- Long branches in Africa, but star-shaped structure outside (characteristic of recent expansion).
- 175,000 ± 50,000 years to common ancestor ( mitochondrial Eve of all people in the study).
- 52,000 ± 28,000 years to branch between the last African and the non-African branch ( mitochondrial Eve of all non-Africans in the study).
- Signal for an expansion in the non-African branch about 1925 generations ago, i.e. about 38,500 years ago if one assumes a generation time of 20 years.
In 2013, another study was finally published in Science , according to which "mitochondrial Eve" lived 99,000 to 148,000 years ago and the so-called Adam of the Y chromosome 120,000 to 156,000 years ago.
Haplotypes
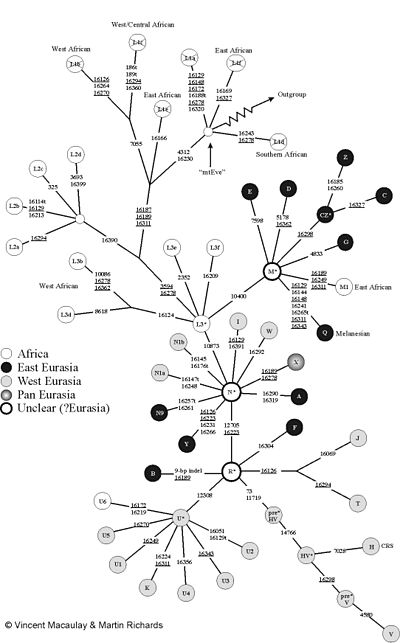
the numbers indicate the position of the mutations.
“MtEve” is the mitochondrial Eve . “Outgroup” leads to mtDNA from other primates (e.g. chimpanzees). The figure uses the usual (incorrect) nomenclature with the "L1 haplogroup": However, L1 forms the root (L1a is no more closely related to L1f than to V!). Therefore the L1 fields have been crossed out.
The mitochondrial DNA of humans can be divided into so-called haplogroups. A haplogroup can itself contain further sub-haplogroups, which in turn can be further subdivided. One tries to map this tree structure with the nomenclature of the haplogroups and alternately uses letters and numbers. Two mtDNAs of a haplogroup are always monophyletic . Characteristic mutations in the gene sequences of the mtDNA outside of the D-loop are used for the assignment .
A person can e.g. B. have the haplogroup C1a3b2. Your mtDNA is then closely related to that of another person, e.g. B. has C1a3b4. Of course, their mtDNA also shares a common ancestor with a third person who has C1a3c5, but that common ancestor had previously lived before the C1a3 lineage split. That is, C1a3b4 and C1a3b2 are monophyletic to C1a3c5. Likewise, C1a3b2 and C1a3c5 are monophyletic towards all H-haplotypes etc.
Unfortunately, the nomenclature has been implemented relatively inconsistently. Many letters have been used to denote the major non-African haplogroups. However, many old haplogroups occur in Africa. These are known collectively as “L” and are already used for subdividing the main groups into digits. There is still no scientific consensus on the assignment of some African haplotypes (in L1 and L3).
If you start from the root, the human mitochondrial family tree consists of a series of deep branches. These genetic lines are now called L1. Unlike earlier thought, L1 is not a monophyletic haplogroup, but forms the root. So L1 are actually a whole package of African haplogroups, which are as old as mitochondrial Eve and whose exact relationship to each other has not yet been clarified.
A branch branches off from these old L1 branches through a mutation at position 10810. The haplogroup L2 in turn splits off from this through a mutation at position 16390. L2 also occurs practically only in sub-Saharan Africans.
A mutation at position 3594 forms the branch on which the large haplogroups M and N as well as numerous other African haplogroups, which are still summarized today under L3, are located. Like L1, L3 is not a true (monophyletic) haplogroup. The haplogroups M and N occur in the vast majority of non-Africans. They are very rare in sub-Saharan Africa, where L1, L2, and L3 dominate.
The haplogroup M is divided into the major haplogroups M1, Z, C, D, E, G and Q. The haplogroup N in N1a, N1b, N9, A, I, W, X and Y, as well as in the haplogroup R, which forms the sub-haplogroups B, F, H, P, T, J, U and K.
The currently most extensive study of mitochondrial DNA was carried out by the Genographic Consortium (see also The Genographic Project ). In this comparison 78,590 genotypic samples were included and the mitochondrial haplogroups (and their subgroups) were represented in a phylogenetic tree .
Geographical distribution
The “old” haplotypes from the L branches dominate in sub-Saharan Africa. There is no doubt that they originated there. These haplotypes are also found in North Africa (approx. 50% frequency) and, to a lesser extent, in Europe and Western Asia.
Haplogroups M and N dominate in the rest of the world and are rare in sub-Saharan Africa. Special variants of the haplogroup M (M1) occur with a frequency of about 20% in Ethiopia. Either M has already arisen there or it is a Semitic return migration to the south.
Native Americans have haplogroups A, B, C, D, and X; of these, A, B, and X emerged from an eastern branch of the haplogroup N, C and D, on the other hand from haplogroup M.
In Europe and Western Asia, haplogroup M is extremely rare. The most common subgroups belong to the subgroup R: H, V, T, J, U and K. In addition, the haplogroups I, W and X occur with a significant frequency. In Europe, the Caucasus and the Middle East, practically the same haplogroups can be found, only the frequencies of the individual haplogroups fluctuate. Haplogroup H in particular is significantly rarer in the Middle East and the Caucasus than in Europe (≈25% versus ≈45%), while haplogroup K is significantly more common. Within Europe, the frequencies of the haplogroups vary slightly depending on the region.
South and East Asia differ greatly from West Asia in terms of haplogroups. The haplogroups C, D, E, G, Z and Q appear here from haplogroup M. The haplogroup N also occurs here, but it is mainly represented by the haplogroups A, B, F, Y and X.
Haplogroup X is noteworthy because it occurs throughout Eurasia and North America, albeit with a relatively low frequency. It used to be assumed that haplogroup X originated in Europe and only occurs in Europe. When the haplogroup was discovered among Native Americans, the hypothesis arose that it came to America from Europe by sea through European emigrants thousands of years ago. In the meantime, however, haplogroup X has also been discovered in Asia (Derneko et al. , 2001).
Ancient mitochondrial DNA
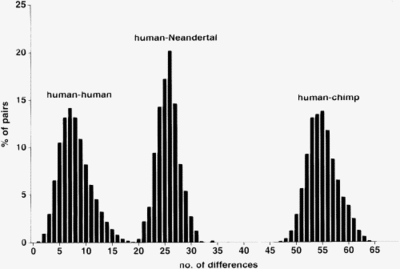
Every cell contains many mitochondria, which in turn contain multiple copies of mitochondrial DNA. This often enables DNA sequencing of mtDNA from fossils that are not too old (less than 100,000 years). By 2006, parts of the mtDNA from twelve Neanderthals from Germany , Croatia , Russia , France , Belgium , Italy and Spain had been sequenced. The result:
- The mtDNA of a Neanderthal man differs about three times as much from that of a modern person as the mtDNA of two modern people from one another.
- The mtDNA of the Neanderthals differ only slightly from one another. The mtDNA of modern people differ significantly from one another.
- The mtDNA of the Neanderthals shows no greater similarity to the mtDNA of today's Europeans than to that of today's Africans or Asians.
The mtDNA of the Neanderthal man split off from the line that led to modern humans relatively early. It is estimated that the common mitochondrial Eve of modern humans and Neanderthals lived between 550,000 and 690,000 years ago, significantly earlier than the mitochondrial Eve of modern humans (Krings et al. , 1997).
According to Nordborg (1998) and others, despite these data, it cannot be ruled out that there was a mixture of Neanderthals and modern humans. On the other hand, Currat & Excoffier (2004) believed that this mtDNA data could practically rule out mixing. Research results from the years 2012 to 2014 on the fossils of Peştera cu oasis in Romania and Ust-Ischim, however, showed hybridizations with Neanderthals.
The mtDNA from all fossil remains of anatomically modern humans ( Homo sapiens ) that have been investigated up to now, however, are closely related to those of humans living today (Serre et al. 2004). The mtDNA from “ Ötzi ”, for example, belonged to the mitochondrial haplogroup K1 (Rollo et al. 2006), more precisely the subtype K1f.
Genetic diversity of mtDNA
The diversity of mitochondrial DNA sequences is greatest in Africa. Overall, the genetic diversity in humans is low compared to the great apes. In a certain segment of mtDNA, chimpanzees show three to four times more genetic diversity than humans. The mtDNAs of 19 examined chimpanzees from the Taï National Park ( Ivory Coast ) alone show a higher diversity than that of all humans, although those monkeys belong to a small, genetically mixed group (Gagneux et al., 1999).
Bonobos , gorillas and orangutans also show a higher genetic diversity of mtDNA than humans (Jobling et al., 2004).
| Locus | Chimpanzee vs. human |
Bonobo vs. human |
Gorilla vs. human |
Orangutan vs. human |
reference | annotation |
|---|---|---|---|---|---|---|
| mtDNA | 3 to 4 times as high | higher | higher | higher | Gagneux, et al. (1999) | - |
| Y chromosome | higher | higher | less | higher | Stone et al. (2002) | see Adam |
| X chromosome | 3 times as high | no data | 2 times as high | 3.5 times as high | Kaessmann et al. (2001) | - |
| Locus | Africa | Asia | Europe | reference | annotation |
|---|---|---|---|---|---|
| mtDNA (pairwise differences) | 2.08 | 1.75 | 1.08 | Vigilant, et al. (1991) | - |
| Y chromosome (43 markers) | 0.841 | 0.904 | 0.852 | Hammer et al. (2001) | see Adam |
| X chromosome (nucleotide diversity) | 0.035 | 0.025 | 0.034 | Kaessmann et al. (1999) | - |
| Autosomes (nucleotide diversity) | 0.115 | 0.061 | 0.064 | Yu et al. (2002) | - |
Eva and the Out of Africa Theory

The theory of mitochondrial Eve is now recognized and established, but there is still no scientific consensus regarding the question of the out-of-Africa theory versus the “ multi-regional origin of modern man ”. It is clear that the theory of mitochondrial Eve does not contradict the Out-of-Africa theory. But it doesn't necessarily refute other models either. After all, it only makes statements about a single locus . The loci in the cell nucleus can have stories that are much more complicated thanks to recombination.
For example, Nordborg (1998) argued that the complete absence of Neanderthal mtDNA in Europeans today could well be the result of genetic drift .
Nordborg (1998) assumed in a simple model that modern man initially spread across Europe without mixing with Neanderthals. It was only after the expansion was complete that modern humans mingled with Neanderthals. According to this model, the Neanderthals could have contributed up to 25% to the gene pool of the mixed population. Due to genetic drift, the mitochondrial lineages of the Neanderthals died out in the meantime ( see also theory ). Today's Europeans could therefore still have DNA of Neanderthal origin in their cell nucleus.
Currat & Excoffier (2004) developed a much more complicated model for the intermingling of modern humans and Neanderthals. The colonization of Europe and the displacement of the Neanderthals by modern humans lasted about 12,000 years (≈500 generations). If the calculations are based on the assumption that expansion and mixing took place at the same time, the Neanderthals get a great advantage because they had colonized Europe before.
If z. B. modern humans, coming from Africa, colonize Anatolia and sometimes mix with the Neanderthals there, the mtDNA of the Neanderthals finds its way into the gene pool of modern humans. If the population of modern humans expands, so does the imported Neanderthal DNA. If now z. For example, if the Balkans are populated by the "mixed offspring" and there is again a mixture with the Neanderthals living there, the proportion of Neanderthal DNA in the gene pool increases even further. The further the wave of expansion of modern humans advances to the west and north, and the more it mixes with the Neanderthals living there, the more Neanderthal DNA accumulates in the gene pool.
According to this model, even a small mixing quota would be sufficient for the mitochondrial DNA of the Neanderthals to completely displace that of modern humans (which was not the case). Currat and Excoffier (2004) found that in the 12,000 years in which modern humans and Neanderthals lived in Europe, there could only be a maximum of 120 "mixed children". Because of this extremely low number, the authors hypothesized that modern humans and Neanderthals are different species that were unable to reproduce together.
Large parts of Asia were already inhabited by members of the genus Homo ( Homo erectus ) when they were colonized by modern people from Africa. Again, there are no mitochondrial lineages that would indicate a mixture.
In summary, it can be said that mitochondrial Eve supports the Out-of-Africa theory with the following facts:
- Mitochondrial DNA in humans shows little genetic diversity (Gagneux et al., 1999).
- The mitochondrial Eve is relatively young at only ≈175,000 years (Ingman et al., 2000).
- The mitochondrial tree shows deep branches in Africa but star-shaped structure outside (Ingman et al., 2000).
- The mitochondrial DNA of Neanderthals is clearly different from that of modern humans (Serre et al., 2004).
- Mixing modern humans with Neanderthals was still considered unlikely until 2004 (Currat & Excoffier, 2004).
Investigations at other loci on the X chromosome, Y chromosome and autosomes also indicate a young, African origin of humans (Takahata et al., 2001).
Between 2013 and 2015, the research team led by the Swedish scientist Svante Pääbo published new findings on mixing :
- Improved analysis methods showed that gene flow took place with a contribution of up to 4% Neanderthal genes to the gene pool of today's Europeans and Asians.
- Research data on the Homo sapiens fossils from Peştera cu oasis in Romania and Ust-Ischim in Siberia supported these statements.
- However, gene flow has so far only been detected in one direction, namely the mating of Homo sapiens men with Neanderthal women.
"Eva" in the popular reception
- Bryan Sykes wrote a book called The Seven Daughters of Eve .
- In River Out of Eden , Richard Dawkins describes the human ancestors in the context of a river of genes and shows that mitochondrial Eve is one of many common ancestors that we can trace back through the various genetic pathways.
- The Discovery Channel aired a documentary called The Real Eve .
- The Japanese novel, horror film and the video game series Parasite Eve use the mitochondrial Eve theory as the basis for a fantasy story about a scientist who resuscitates his wife with the help of regenerated cells, which has fatal consequences.
- Greg Egan wrote a short story called Mitochondrial Eve .
- In Ronald D. Moore's science fiction series Battlestar Galactica , Hera, the daughter of a human and a Cylon, is the mitochondrial Eve .
- Lynn Okamoto used the theory of mitochondrial Eve for his manga Elfen Lied . In this one, Lucy is the mitochondrial Eve of a new breed called Diclonius.
See also
literature
- Specialist literature
- M. Currat, L. Excoffier: Modern humans did not admix with Neanderthals during their range expansion into Europe. In: PLoS Biology . Lawrence 2.2004,12, e421. doi : 10.1371 / journal.pbio.0020421 ISSN 1544-9173 .
- MV Derenko, T. Grzybowski, BA Malyarchuk, J. Czarny, DM Sliwka, IA Zakharov: The presence of mitochondrial haplogroup x in Altaians from South Siberia. In: American Journal of Human Genetics (Am J Hum Genet). New York 69.2001,1, 237-241. PMID 11410843 ISSN 0002-9297 .
- P. Gagneux, C. Wills, U. Gerloff, D. Tautz, PA Morin, C. Boesch, B. Fruth, G. Hohmann, OA Ryder, DS Woodruff: Mitochondrial sequences show diverse evolutionary histories of African hominoids. In: Proceedings of the National Academy of Sciences of the United States of America (PNaS USA). Washington 96.1999,9, 5077-5082. doi : 10.1073 / pnas.96.9.5077 ISSN 0027-8424 .
- MF Hammer, TM Karafet, AJ Redd, H. Jarjanazi, S. Santachiara-Benerecetti, H. Soodyall, SL Zegura: Hierarchical patterns of global human Y-chromosome diversity. In: Molecular biology and evolution (Mol Biol Evol). Oxford 18.2001,7, 1189-1203. PMID 11420360 ISSN 0737-4038 .
- M. Ingman, H. Kaessmann, S. Pääbo, U. Gyllensten: Mitochondrial genome variation and the origin of modern humans. In: Nature . London 408.2000,6813, 708-713. doi : 10.1038 / 35047064 ISSN 0028-0836 .
- Mark A. Jobling, Chris Tyler-Smith, Matthew Hurles: Human Evolutionary Genetics. Origins, Peoples and Disease. ISBN 0-8153-4185-7 .
- H. Kaessmann, F. Heissig, A. von Haeseler, S. Pääbo: DNA sequence variation in a non-coding region of low recombination on the human X chromosome. In: Nature Genetics (Nat Genet). New York 22.1999, 1, 78-81. doi : 10.1038 / 8785 ISSN 1061-4036 .
- H. Kaessmann, V. Wiebe, G. Weiss, S. Pääbo: Great ape DNA sequences reveal a reduced diversity and an expansion in humans. In: Nature Genetics (Nat Genet). New York 27.2001,2, 155-156. doi : 10.1038 / 84773 ISSN 1061-4036 .
- M. Krings, A. Stone, RW Schmitz, H. Krainitzki, M. Stoneking, S. Pääbo: Neandertal DNA sequences and the origin of modern humans. In: Cell. Cambridge 90.1997,1, 19-30. doi : 10.1016 / S0092-8674 (00) 80310-4 ISSN 0092-8674 .
- M. Nordborg: On the probability of Neanderthal ancestry. In: American Journal of Human Genetics (Am J Hum Genet). New York 63.1998,4, 1237-1240. PMID 9758610 ISSN 0002-9297 .
- F. Rollo, L. Ermini, S. Luciani, I. Marota, C. Olivieri, D. Luiselli: Fine characterization of the Iceman's mtDNA haplogroup. in: American journal of physical anthropology (Am J Phys Anthropol). New York 130.2006,4, 557-564. doi : 10.1002 / ajpa.20384 ISSN 0002-9483 .
- Serre, D .; Langaney, A .; Chech, M .; Nicola, MT; Paunovic, M .; Mennecier, P .; Hofreiter, M .; Possnert, G. & Pääbo, S. (2004): No evidence of Neandertal mtDNA contribution to early modern humans. PLoS Biol 2 (3), E57. doi : 10.1371 / journal.pbio.0020057 .
- Stone, AC; Griffiths, RC; Zegura, SL & Hammer, MF (2002): High levels of Y-chromosome nucleotide diversity in the genus Pan. Proc Natl Acad Sci USA 99 (1), 43-48. doi : 10.1073 / pnas.012364999 .
- Vigilant, L .; Stoneking, M .; Harpending, H .; Hawkes, K. & Wilson, AC (1991): African populations and the evolution of human mitochondrial DNA. Science 253 (5027): 1503-1507. doi : 10.1126 / science.1840702 .
- Takahata, N .; Lee, SH & Satta, Y. (2001): Testing multiregionality of modern human origins. Mol Biol Evol 18 (2), 172-183. PMID 11158376 .
- Yu, N .; Chen, F .; Ota, S .; Jorde, LB; Pamilo, P .; Patthy, L .; Ramsay, M .; Jenkins, T .; Shyue, S. & Li, W. (2002): Larger genetic differences within africans than between Africans and Eurasians. Genetics 161 (1), 269-274. PMID 12019240 .
- reception
- Bryan Sykes: The Seven Daughters of Eve. Lübbe-Verlag, Bergisch Gladbach 2001, ISBN 3-7857-2060-2 .
Web links
- Geneticancestor: Mitochondrial Eva
- Mathematical consideration by Franz Embacher
- Spread of the Y-Haplogrouppen (PDF file; 386 kB)
- Spread based on Roots for Real's largest genetic database
Individual evidence
- ↑ cf. J. Hein, MH Schierup, C. Wiuf: Gene genealogies, variation and evolution: a primer in coalescent theory. Oxford University Press, Oxford 2005.
- ^ Alan R. Templeton: Haplotype trees and modern human origins. Yearbook of physical Anthropology 48, 2005, pp. 33-59.
- ↑ CF Aquadro, BD Greenberg: Human mitochondrial DNA variation and evolution, analysis of nucleotide sequences from seven individuals. (PDF; 1.7 MB) In: Genetics. Vol. 103, Bethesda 1983, pp. 287-312. PMID 6299878 ISSN 0016-6731
- ↑ MJ Johnson et al. a .: Radiation of human mitochondria DNA types analyzed by restriction endonuclease cleavage patterns. In: Journal of molecular evolution. Vol. 19, New York 1983, pp. 255-271. PMID 6310133 doi : 10.1007 / BF02099973 ISSN 0022-2844
- ↑ RL Cann et al.: Mitochondrial DNA and human evolution. In: Nature . Vol. 325, London 1987, pp. 31-36. PMID 3025745 doi : 10.1038 / 325031a0 ISSN 0028-0836
- ↑ G. David Poznik et al .: Sequencing Y Chromosomes Resolves Discrepancy in Time to Common Ancestor of Males Versus Females. In: Science . Volume 341, No. 6145, 2013, pp. 562-565, doi: 10.1126 / science.1237619
- ↑ Macaulay and Richards
- ↑ DM Behar u. a .: The Genographic Project public participation mitochondrial DNA database. In: PLoS Genet . Vol. 3, San Francisco 2007, pp. E104. PMID 17604454 doi : 10.1371 / journal.pgen.0030104 ISSN 1553-7390
- ↑ RE Green u. a .: Analysis of one million base pairs of Neanderthal DNA. In: Nature. Vol. 444, London 2006, pp. 330-336. PMID 17108958 doi : 10.1038 / nature05336 ISSN 0028-0836
- ↑ a b Early Europeans mixed with Neanderthals. Retrieved from mpg.de on July 12, 2015, with an image of the lower jaw Oase1
- ↑ a b Genome of the oldest modern human being deciphered. Max Planck Society of October 22, 2014.
- ↑ European Academy of Bozen / Bolzano (EURAC), January 15, 2016 - NPO
- ↑ Gene flow from Neanderthals to Homo sapiens. Retrieved from mpg.de on June 28, 2015.
- ↑ Neanderthal genome detected in Homo sapiens, no detection of sapiens genome in Neanderthals yet. Retrieved from mpg.de on July 20, 2015.