Immunoreceptor tyrosine-based activation motif
An immunoreceptor tyrosine-based activation motif (ITAM) is a conserved sequence of four amino acids that is repeated twice in the cytoplasmic tails of Non-catalytic tyrosine-phosphorylated receptors, cell-surface proteins found on immune cells.[1] It is an integral component for the initiation of the signaling pathway and subsequently the activation of immune cells.
Structure
The motif contains a tyrosine separated from a leucine or isoleucine by any two other amino acids, giving the signature YxxL/I.[1] Two of these signatures are typically separated by between 6 and 8 amino acids in the cytoplasmic tail of the molecule (YxxL/Ix(6-8)YxxL/I).
Function
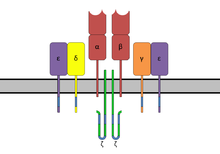
ITAMs are important for signal transduction in immune cells. They are found in the cytoplasmic tails of non-catalytic tyrosine-phosphorylated receptors [2] such as the CD3 and ζ-chains of the T cell receptor complex,[1] the CD79-alpha and -beta chains of the B cell receptor complex, and certain Fc receptors.[2] The tyrosine residues within these motifs become phosphorylated by Src family kinases (SFK) following interaction of the receptor molecules with their ligands. Phosphorylated ITAMs serve as docking sites for other proteins containing a SH2 domain inducing a signaling cascade resulting in the activation of the immune cell.
Other non-catalytic tyrosine-phosphorylated receptors carry a conserved inhibitory motif (ITIM) that, when phosphorylated, results in the inhibition of the signaling pathway.
Genetic Variations
Rare human genetic mutations are catalogued in the human genetic variation databases[3][4][5] which can reportedly result in creation or deletion of ITIM and ITAMs[6].
References
- ^ a b c Abbas, Abul K; Lichtman, Andrew H. (2009), Basic Immunology: Functions and Disorders of the Immune System (3 ed.), Philadelphia, PA: Saunders, ISBN 978-1-4160-4688-2
{{citation}}: Unknown parameter|name-list-format=ignored (|name-list-style=suggested) (help) - ^ a b Dushek O, Goyette J, van der Merwe PA (November 2012). "Non-catalytic tyrosine-phosphorylated receptors". Immunological Reviews. 250 (1): 258–76. doi:10.1111/imr.12008. PMID 23046135.
- ^ Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, et al. (October 2015). "A global reference for human genetic variation". Nature. 526 (7571): 68–74. Bibcode:2015Natur.526...68T. doi:10.1038/nature15393. PMC 4750478. PMID 26432245.
- ^ Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K (January 2001). "dbSNP: the NCBI database of genetic variation". Nucleic Acids Research. 29 (1): 308–11. doi:10.1093/nar/29.1.308. PMC 29783. PMID 11125122.
- ^ Cummings BB, Karczewski KJ, Kosmicki JA, Seaby EG, Watts NA, Singer-Berk M, et al. (May 2020). "Transcript expression-aware annotation improves rare variant interpretation". Nature. 581 (7809): 452–458. Bibcode:2020Natur.581..452C. doi:10.1038/s41586-020-2329-2. PMC 7334198. PMID 32461655.
- ^ Ulaganathan VK (May 2020). "TraPS-VarI: Identifying genetic variants altering phosphotyrosine based signalling motifs". Scientific Reports. 10 (1): 8453. Bibcode:2020NatSR..10.8453U. doi:10.1038/s41598-020-65146-2. PMC 7242328. PMID 32439998.
