Gibson Assembly
Gibson assembly (also Gibson isothermal assembly , German: isothermal assembly according to Gibson ) is a biochemical method for the generation and replication of DNA . It is a variant of isothermal DNA amplification . In combination with a partial exonuclease digest, it is used for cloning .
principle
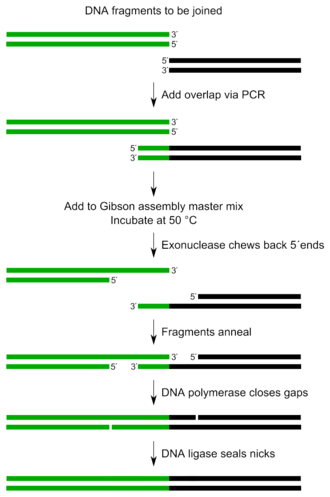
The Gibson assembly takes place at a constant temperature ( isothermal ). The DNA double strands to be joined together must have overlapping DNA sequences in an isothermal assembly . The T5 exonuclease partially digests the 5 'ends of a DNA double strand, as a result of which the overhanging second strands, as sticky ends, initiate a hybridization of the homologous single-stranded regions of the DNA sequences. Furthermore, a DNA polymerase with DNA double-strand separating properties is used, which separates the double strand present during the synthesis of a second DNA strand and displaces the existing second strand. The φ29 DNA polymerase or the large fragment of the Bst DNA polymerase from Bacillus stearothermophilus , which is thermostable at 65 ° C. , can be used here. A ligase can be used to link the generated DNA sequences together in vitro .
Compared to the different variants of the polymerase chain reaction , this method is particularly suitable for the replication of long DNA sequences (more than 2,000 to 100,000 base pairs ). Another advantage is that with mixtures of DNA there is no bias due to this type of amplification . This means that in the case of mixtures, individual DNA sequences are not preferred or others are disadvantaged. Alternative isothermal methods for replicating DNA are e.g. B. multidisplacement amplification , recombinase polymerase amplification , loop-mediated isothermal amplification (LAMP), helicase-dependent amplification (HDA) and the nicking enzyme amplification reaction (NEAR). Alternative methods for the targeted generation of new DNA sequences are in addition to the three methods described by Gibson z. B. fusion PCR, polymerase cycling assembly (PCA), circular polymerase extension cloning (CPEC) or homologous recombination in vivo .
Applications
The Gibson Assembly is used, among other things, for cloning and in synthetic biology for the amplification of DNA.
Individual evidence
- ^ DG Gibson, L. Young, RY Chuang, JC Venter, CA Hutchison, HO Smith: Enzymatic assembly of DNA molecules up to several hundred kilobases. In: Nature methods. Volume 6, Number 5, May 2009, ISSN 1548-7105 , pp. 343-345, doi : 10.1038 / nmeth.1318 , PMID 19363495 .
- ^ DG Gibson, HO Smith, CA Hutchison, JC Venter, C. Merryman: Chemical synthesis of the mouse mitochondrial genome. In: Nature methods. Volume 7, Number 11, November 2010, ISSN 1548-7105 , pp. 901-903, doi : 10.1038 / nmeth.1515 , PMID 20935651 .
- ↑ a b D. G. Gibson: Enzymatic assembly of overlapping DNA fragments. In: Methods in enzymology. Volume 498, 2011, ISSN 1557-7988 , pp. 349-361, doi : 10.1016 / B978-0-12-385120-8.00015-2 , PMID 21601685 .
- ^ WP Stemmer, A. Crameri, KD Ha, TM Brennan, HL Heyneker: Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. In: Genes. Volume 164, Number 1, October 1995, ISSN 0378-1119 , pp. 49-53, PMID 7590320 .
- ^ HO Smith, CA Hutchison, C. Pfannkoch, JC Venter: Generating a synthetic genome by whole genome assembly: phiX174 bacteriophage from synthetic oligonucleotides. In: Proceedings of the National Academy of Sciences . Volume 100, number 26, December 2003, ISSN 0027-8424 , pp. 15440-15445, doi : 10.1073 / pnas.2237126100 , PMID 14657399 , PMC 307586 (free full text).
- ↑ J. Quan, J. Tian: Circular polymerase extension cloning of complex gene libraries and pathways. In: PloS one. Volume 4, number 7, 2009, p. E6441, ISSN 1932-6203 , doi : 10.1371 / journal.pone.0006441 , PMID 19649325 , PMC 2713398 (free full text).
- ^ T. Ellis, T. Adie, GS Baldwin: DNA assembly for synthetic biology: from parts to pathways and beyond. In: Integrative biology: quantitative biosciences from nano to macro. Volume 3, Number 2, February 2011, ISSN 1757-9708 , pp. 109-118, doi : 10.1039 / c0ib00070a , PMID 21246151 .