Deoxyribonucleic acid

deoxyribonucleic acid (abbreviated DNS ), in German now mostly referred to as DNA (abbreviation for English deoxyribonucleic acid ), is a nucleic acid that is composed of a chain of many nucleotides as a polynucleotide . The biomolecule in the chromosomes is the carrier of genetic information in all living things and in many viruses ( DNA viruses , pararetroviruses ) , i.e. the material basis of the genes. The word is made up of des- (English: de-), the first two syllables of Oxygenium (oxygen), the first two syllables of ribose (see deoxyribose ) and nucleic acid .
In its normal state, DNA is built up in the form of a double helix . Its building blocks are four different nucleotides, each consisting of a phosphate residue , the sugar deoxyribose and one of four organic bases ( adenine , thymine , guanine and cytosine , often abbreviated as A, T, G and C).
The genes in the DNA contain the information for the production of ribonucleic acids (RNA, in German also RNS). For protein-coding genes, this is an important RNA group, the mRNA ( English messenger RNA ). In turn, it contains the information for the construction of proteins , which are necessary for the biological development of a living being and the metabolism in the cell . The sequence of the bases determines the sequence of the amino acids of the respective protein: the genetic code encodes a specific amino acid with three neighboring bases .
In the cells of eukaryotes , which also include plants , animals and fungi , most of the DNA in the nucleus (from the Latin nucleus , hence nuclear DNA or nDNA) is organized as chromosomes . A small part is located in the mitochondria , the "power plants" of the cells, and is accordingly called mitochondrial DNA (mtDNA). Plants and algae are also DNA in photosynthesis operated organelles , the chloroplasts or plastids ( cpDNA ). In bacteria and archaea - the prokaryotes that do not have a nucleus - the DNA is located in the cytoplasm . Some viruses , so-called RNA viruses , store their genetic information in RNA instead of DNA.
Discovery story
In 1869 the Swiss physician discovered Friedrich Miescher in an extract from pus one by mild acid treatment from the nuclei of leukocytes recovered substance he nucleic called. Back then, Miescher was working in Felix Hoppe-Seyler's laboratory in Tübingen Castle . In 1889 the German Richard Altmann isolated proteins and nucleic acids from the nucleus. Further findings on nucleic acids can be traced back to the work of Albrecht Kossel (see “ The discovery of nucleic bases ”), for whom he was awarded the Nobel Prize in Physiology or Medicine in 1910 . In 1885 he announced that a nitrogen-rich base with the molecular formula C 5 H 5 N 5 had been isolated from a large amount of bovine pancreas , for which he suggested the name adenine , derived from the Greek word “aden” for gland . In 1891 Kossel was able to produce yeast nucleic acid (using Altmann's method) and detect adenine and guanine as cleavage products. It turned out that a carbohydrate also had to be part of the nucleic acid. Kossel chose the name nucleic bases for the basic substances guanine and adenine and their derivatives .
In 1893 Kossel reported that he had extracted nucleic acid from the calf's thymus glands and obtained a well-crystallized cleavage product, for which he proposed the name thymine . In 1894 he isolated another (basic) substance from the thymus glands. Kossel named it cytosine .
After the structural formulas of guanine and adenine as the purine body and thymine as the pyrimidine body had been finally elucidated at the end of the 19th century - mainly through the syntheses of Emil Fischer - Kossel and Hermann Steudel (1871–1967) were also able to develop the structural formula of the nucleic base cytosine determine beyond doubt as a pyrimidine body. It had meanwhile been shown that guanine, adenine, thymine and cytosine can be found in all viable cells.
The knowledge about these four nucleobases should be of essential importance for the later structure elucidation of the DNA. It was Albrecht Kossel who - together with a carbohydrate and phosphoric acid - clearly characterized them as building blocks of nucleic acid:
“I managed to get a number of fragments ... which are characterized by a very peculiar collection of nitrogen atoms. There are next to each other ... the cytosine, the thymine, the adenine and the guanine. ”(Nobel lecture on December 12, 1910).
The biochemist Phoebus Levene from Lithuania proposed a chain-like structure of the nucleic acid in which the nucleotides are joined together by the phosphate residues and repeat themselves. In 1929, in collaboration with the Russian physiologist Efim London (1869-1932), he was able to identify the sugar content of "animal nucleic acid" as deoxyribose (J. Biol. Chem. 1929, 83. pages 793-802). [5a] Only subsequently was it referred to as deoxyribonucleic acid. It was recognized that it also occurs in plant cell nuclei.
As early as 1932, K. Voit and Hartwig Kuhlenbeck regarded DNA as an effective component of chromosomes or nuclear chromatin . In 1937 William Astbury published X-ray diffraction patterns for the first time , which indicated a repetitive structure of DNA.
In 1943, Oswald Avery demonstrated that the transformation of bacteria, i.e. the transfer of hereditary information from one bacterial strain to another (now called horizontal gene transfer ), is based on the transfer of DNA. This contradicted the generally favored assumption at the time that it was not DNA but proteins that were the carriers of genetic information. Avery received support in his interpretation in 1952 when Alfred Hershey and Martha Chase demonstrated that DNA contains the genetic information of the T2 phage .
The American James Watson and the British Francis Crick succeeded in decoding the structure of DNA and recreating it in a model on February 28, 1953. They published their discovery in the April 1953 issue of Nature magazine in their famous article Molecular Structure of Nucleic Acids: A Structure for Deoxyribose Nucleic Acid . Watson came to England in 1951 after completing his PhD a year earlier at Indiana University Bloomington in the United States. Although he had received a scholarship in molecular biology , he was increasingly concerned with the question of human genome. Crick was doing his doctorate on the crystal structure of the hemoglobin molecule in Cambridge, unsuccessfully , when he met Watson in 1951.
At that time, a bitter race for the structure of DNA had already broken out, in which Linus Pauling at the California Institute of Technology (Caltech) was participating. Watson and Crick had actually been assigned to other projects and had no significant expertise in chemistry . They based their thinking on the research of the other scientists.
Watson said he wanted to decipher the genome without having to learn chemistry. In a conversation with the renowned chemist and creator of the Chargaff rules , Erwin Chargaff , Crick forgot important molecular structures, and Watson made inappropriate comments in the same conversation that revealed his ignorance of the field of chemistry. Chargaff then called the young colleagues "scientific clowns".
Watson visited in late 1952 at King's College , London, Maurice Wilkins , who showed him DNA x-rays of Rosalind Franklin (which happened against Franklin's will). Watson saw at once that it is the molecule is a double - helix had to act; Franklin himself had suspected the presence of a helix based on the data, but she had no convincing model for the structure to show. Since it was known that the purine and pyrimidine bases form pairs, Watson and Crick were able to deduce the entire molecular structure. For example, at the Cavendish Laboratory at Cambridge University , they developed the double helix model of DNA with the base pairs in the middle, which was published in the journal Nature on April 25, 1953.
Towards the end of this memorable publication, the sentence "It has not escaped our notice that the specific pairing we have postulated immediately suggests a possible copying mechanism for the genetic material" . ("It has not escaped our attention that the specific pairing that we take for granted suggests a possible mechanism of replication for the genetic material.")
“For their discoveries about the molecular structure of nucleic acids and their importance for the transmission of information in living matter”, Watson and Crick received the 1962 Nobel Prize for Medicine together with Maurice Wilkins .
Rosalind Franklin , whose X-ray diffraction diagrams had contributed significantly to the decoding of the DNA structure, had already died at this point and could therefore no longer be nominated.
For further historical information on deciphering the inheritance processes see “ Research History of the Cell Nucleus ” as well as “ Research History of Chromosomes ” and “ Chromosome Theory of Inheritance ”.
Structure and organization
building blocks
Deoxyribonucleic acid is a long chain molecule ( polymer ) made up of many building blocks called deoxyribonucleotides or nucleotides for short . Every nucleotide has three components: phosphoric acid or phosphate, the sugar deoxyribose and a heterocyclic nucleobase or base for short. The deoxyribose and phosphoric acid subunits are the same for each nucleotide. They form the backbone of the molecule. Units made up of base and sugar (without phosphate) are known as nucleosides .
The phosphate residues are hydrophilic due to their negative charge , they give DNA in aqueous solution an overall negative charge. Since this negatively charged DNA dissolved in water cannot release any further protons , strictly speaking it is not (any longer) an acid . The term deoxyribonucleic acid refers to an uncharged state in which protons are attached to the phosphate residues.
The base can be a purine , namely adenine ( A ) or guanine ( G ), or a pyrimidine , namely thymine ( T ) or cytosine ( C ). Since the four different nucleotides only differ in their base, the abbreviations A, G, T and C are also used for the corresponding nucleotides.
The five carbon atoms of a deoxyribose are numbered from 1 '(pronounced one line ) to 5'. The base is attached to the 1 'end of this sugar. The phosphate residue hangs at the 5 'end. Strictly speaking, deoxyribose is 2-deoxyribose; The name comes from the fact that, compared to a ribose molecule, an alcoholic hydroxyl group (OH group) is missing at the 2'-position (i.e. replaced by a hydrogen atom).
At the 3 'position there is an OH group which links the deoxyribose to the 5' carbon atom of the sugar of the next nucleotide via a so-called phosphodiester bond (see figure). As a result, each so-called single strand has two different ends: a 5 'and a 3' end. DNA polymerases , which synthesize DNA strands in the living world, can only add new nucleotides to the OH group at the 3 'end, but not at the 5' end. The single strand always grows from 5 'to 3' (see also DNA replication below). A nucleoside triphosphate (with three phosphate residues) is delivered as a new component, from which two phosphates are split off in the form of pyrophosphate . The remaining phosphate residue of the newly added nucleotide is connected to the OH group at the 3 'end of the last nucleotide present in the strand with elimination of water. The sequence of bases in the strand encodes the genetic information.
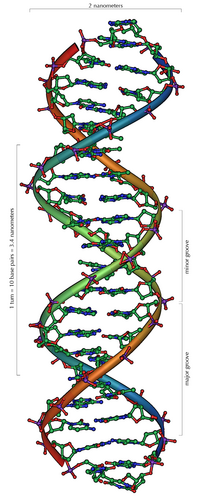
The double helix

DNA usually occurs as a helical double helix in a conformation before, the B-DNA is called. Two of the single strands described above are attached to one another, namely in opposite directions: at each end of the double helix, one of the two single strands has its 3 'end, the other its 5' end. Due to the juxtaposition, there are always two specific bases facing each other in the middle of the double helix, they are "paired". The double helix is mainly stabilized by stacking interactions between successive bases on the same strand (and not, as is often claimed, by hydrogen bonds between the strands).
Adenine and thymine are always paired, which form two hydrogen bonds, or cytosine with guanine, which are connected to one another via three hydrogen bonds. A bridge is formed between the molecular positions 1═1 and 6═6, with guanine-cytosine pairings additionally between 2 zusätzlich2. Since the same bases are always paired, the sequence of the bases in one strand can be used to derive that of the other strand; the sequences are complementary (see also: base pair ). The hydrogen bonds are almost exclusively responsible for the specificity of the pairing, but not for the stability of the double helix.
Since a purine is always combined with a pyrimidine, the distance between the strands is the same everywhere, resulting in a regular structure. The whole helix has a diameter of about 2 nm and winds 0.34 nm further with each sugar molecule.
The planes of the sugar molecules are at an angle of 36 ° to each other, and a complete rotation is consequently achieved after 10 bases (360 °) and 3.4 nm. DNA molecules can get very large. For example, the largest human chromosome contains 247 million base pairs.
When the two individual strands are wound around each other, lateral gaps remain so that the bases here lie directly on the surface. There are two of these furrows that wind around the double helix (see illustrations and animation at the beginning of the article). The "large furrow" is 2.2 nm wide, the "small furrow" only 1.2 nm.
Accordingly, the bases in the major furrow are more accessible. Proteins that bind to the DNA in a sequence-specific manner, such as transcription factors , therefore usually bind to the major groove.
Some DNA dyes, such as DAPI , also attach to a groove.
The accumulated binding energy between the two individual strands holds them together. There are no covalent bonds here, so the DNA double helix does not consist of one molecule, but of two. This allows the two strands to be temporarily separated in biological processes.
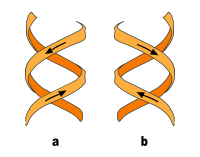
In addition to the B-DNA just described, there is also A-DNA as well as a left-handed, so-called Z-DNA, which was first examined by Alexander Rich and his colleagues at MIT in 1979 . This occurs particularly in GC-rich sections. It was not until 2005 that a crystal structure was reported which shows Z-DNA directly in connection with B-DNA and thus provides indications of the biological activity of Z-DNA. The following table and the illustration next to it show the differences between the three forms in direct comparison.
| Structural feature | A-DNA | B-DNA | Z-DNA |
|---|---|---|---|
| Construction from | Monomers | Monomers | Dimers |
| Direction of rotation of the helix | right | right | Left |
| Diameter (approx.) | 2.6 nm | 2.37 nm | 1.8 nm |
| Helical turn per base pair (twist) | 32.7 ° | 34.3 ° | 30 ° |
| Base pairs per helical turn | 11 | 10 | 12 |
| Increase per base | 0.29 nm | 0.34 nm | 0.37 nm |
| Increase per turn (pitch) | 3.4 nm | 3.4 nm | 4.4 nm |
| Inclination angle of the base pairs to the axis | 20 ° | 6 ° | 7 ° |
| Big furrow | tight and deep |
wide and deep depth: 0.85 nm |
flat |
| Small furrow | wide and flat |
narrow and deep depth: 0.75 nm |
tight and deep |
|
Pyrimidine bases (cytosine / thymine / uracil) sugar conformation glycosidic bond |
C3'- endo anti |
C2'- endo anti |
C 2 ' - endo anti |
|
Purine bases (adenine / guanine) sugar conformation glycosidic bond |
C3'- endo anti |
C2'- endo anti |
C 3 ' - endo syn |
The stacks of base pairs ( base stackings ) are not exactly parallel to each other like books, but form wedges which the helix inclined in one or the other direction. The largest wedge is formed by adenosines, which are paired with thymidines on the other strand. As a result, a series of AT pairs form an arc in the helix. If such series follow one another at short intervals, the DNA molecule assumes a bent or curved structure, which is stable. This is also called sequence-induced diffraction, since the diffraction can also be caused by proteins (the so-called protein-induced diffraction). Sequence-induced diffraction is often found at important locations in the genome.
Chromatin and chromosomes
The DNA in the eukaryotic cell is organized in the form of chromatin threads, called chromosomes, which are located in the cell nucleus . A single chromosome contains a long, continuous double strand of DNA (in a chromatid ) from the anaphase to the beginning of the S phase . At the end of the S phase , the chromosome consists of two identical strands of DNA (in two chromatids).
Since such a DNA thread can be several centimeters long, but a cell nucleus is only a few micrometers in diameter, the DNA must also be compressed or “packed”. In eukaryotes, this happens with so-called chromatin proteins, of which the basic histones deserve special mention. They make up the nucleosomes around which the DNA is wrapped at the lowest level of packaging. During nuclear division (mitosis) , each chromosome is condensed into its maximally compact form. This allows them to be identified particularly well in the metaphase with the light microscope .
Bacterial and Viral DNA
In prokaryotic cells, the majority of double-stranded DNA in the cases documented so far is not present as linear strands with a beginning and an end, but rather as circular molecules - each molecule (i.e. each DNA strand) closes with its 3 'and 5 '-End to the circle. These two circular, closed DNA molecules are called a bacterial chromosome or a plasmid , depending on the length of the sequence . In bacteria, they are not located in a cell nucleus, but are freely present in the plasma. The prokaryote DNA is wound up with the help of enzymes (for example topoisomerases and gyrases ) into simple " supercoils " that resemble a coiled telephone cord. By turning the helices around themselves, the space required for the genetic information is reduced. In the bacteria, topoisomerases ensure that by constantly cutting and reconnecting the DNA, the twisted double strand is untwisted at a desired point (prerequisite for transcription and replication ). Viruses contain either DNA or RNA as genetic information, depending on their type. In both the DNA and RNA viruses, the nucleic acid is protected by a protein envelope.
Chemical and physical properties of the DNA double helix
At neutral pH, DNA is a negatively charged molecule, with the negative charges on the phosphates in the backbone of the strands. Although two of the three acidic OH groups of the phosphates are esterified with the neighboring deoxyriboses , the third is still present and emits a proton at a neutral pH value , which causes the negative charge. This property is used in agarose gel electrophoresis to separate different DNA strands according to their length. Some physical properties such as the free energy and the melting point of the DNA are directly related to the GC content , i.e. they are sequence dependent.
Stack interactions
Two factors are mainly responsible for the stability of the double helix: the base pairing between complementary bases and stacking interactions between successive bases.
Contrary to what was initially assumed, the energy gain through hydrogen bonds is negligible, since the bases can form hydrogen bonds of similar quality with the surrounding water. The hydrogen bonds of a GC base pair contribute only minimally to the stability of the double helix, while those of an AT base pair even have a destabilizing effect. Stack interactions, on the other hand, only work in the double helix between successive base pairs: a dipole-induced dipole interaction occurs between the aromatic ring systems of the heterocyclic bases , which is energetically favorable. Thus, the formation of the first base pair is quite unfavorable due to the low energy gain and loss , but the elongation (lengthening) of the helix is energetically favorable since the base pair stacking proceeds with energy gain.
However, the stacking interactions are sequence-dependent and energetically most favorable for stacked GC-GC, less favorable for stacked AT-AT. The differences in the stacking interactions mainly explain why GC-rich DNA segments are thermodynamically more stable than AT-rich ones, while hydrogen bonding plays a subordinate role.
Melting point
The melting point of DNA is the temperature at which the binding forces between the two individual strands are overcome and these separate from each other. This is also known as denaturation .
As long as the DNA denatures in a cooperative transition (which takes place in a narrow temperature range), the melting point describes the temperature at which half of the double strands are denatured into single strands. The correct terms “midpoint of transition temperature” or midpoint temperature T m are derived from this definition .
The melting point depends on the respective base sequence in the helix. It increases if there are more GC base pairs in it, since these are entropically more favorable than AT base pairs. This is not so much due to the different number of hydrogen bonds that the two pairs form, but much more to the different stacking interactions. The stacking energy of two base pairs is much smaller when one of the two pairs is an AT base pair. GC stacks, on the other hand, are energetically more favorable and stabilize the double helix more strongly. The ratio of the GC base pairs to the total number of all base pairs is given by the GC content .
Since single-stranded DNA absorbs about 40 percent more UV light than double-stranded DNA, the transition temperature can be easily determined in a photometer .
When the temperature of the solution falls below T m , the single strands can attach themselves again. This process is called renaturation or hybridization. The interplay between denaturation and renaturation is used in many biotechnological processes, for example in the polymerase chain reaction (PCR), in Southern blots and in in situ hybridization .
Cross-shaped DNA on palindromes
A palindrome is a sequence of nucleotides in which the two complementary strands can be read from the right as well as from the left.
Under natural conditions (with high torsional tension in the DNA) or artificially in a test tube, this linear helix can develop as a cruciform, in which two branches emerge that protrude from the linear double strand. The branches each represent a helix, but at least three nucleotides remain unpaired at the end of a branch. When changing from the cross shape to the linear helix, the base pairing is retained because of the flexibility of the phosphodiester-sugar backbone.
The spontaneous agglomeration of complementary bases to form so-called stem-loop structures is also often observed with single-stranded DNA or RNA.
Non-standard bases
Occasionally, deviations from the above four canonical bases (standard bases) adenine (A), guanine (G), thymine (T) and cytosine (C) are observed in viruses and cellular organisms; further deviations can be generated artificially.
Natural non-standard bases
-
Uracil (U) is normally not found in DNA, it only occurs as a breakdown product of cytosine. However, in several bacteriophages (bacterial viruses ), uracil replaces thymine:
- Bacillus subtilis bacteriophage PBS1 ( ICTV : Species Bacillus virus PBS1 ) and 'PBS2' (proposed species ' Bacillus phage PBS2 ' aka ' Bacteriophage PBS2 ') - both species are myophages, i. H. Phage from the Myoviridae family(with no genus assigned).
- Bacillus virus PBS1 (ICTV: species Yersinia virus R1RT in the genus Tg1virus , family Myoviridae )
-
Staphylococcus -Phage S6 (alias Staphylococcus aureus Bacteriophage 15, also from the Myoviridae family)
Uracil is also found in the DNA of eukaryotes such as Plasmodium falciparum ( Apicomplexa ). It is there in relatively small amounts (7-10 uracil units per million bases).
- 5-Hydroxymethyldeoxyuridine (hm5dU) replaces thymidine in the genome of various Bacillus phages of the species Bacillus virus SPO1 , genus Spo1virus (formerly Spounalikevirus or SPO1-like viruses ), also family Myoviridae . These are the phages SPO1, SP8, SP82, 'Phi-E' alias 'ϕe' and '2C')
- 5-dihydroxypentauracil (DHPU, with nucleotide 5-dihydroxypentyl-dUMP, DHPdUMP) has been described as a replacement for thymidine in the ' Bacillus Phage SP15' (also 'SP-15', family Myoviridae ).
- Beta-d-glucopyranosyloxymethyluracil (base J), also a modified form of uracil, was found in various organisms: the flagellates Diplonema and Euglena (both Excavata : Euglenozoa ) and all genus of the kinetoplastids . The biosynthesis of J is performed in two steps: In the first step, a specific thymidine into DNA in Hydroxymethyldesoxyuridin (HOMedU) is converted, in the second HOMedU becomes J glycosylated . There are some proteins that specifically bind to this base. These proteins appear to be distant relatives of the Tet1 oncogene , which is involved in the pathogenesis of acute myeloid leukemia . J appears to act as a termination signal for RNA polymerase II .
- In 1976 it was found that the 'Cyanophage S-2L' (species ' Cyanobacteria phage S-2L ', genus ' Cyanostylovirus ', family Siphoviridae , formerly Cyanostyloviridae or Styloviridae ), whose hosts are species of the genus Synechocystis , all have adenosine bases in its genome 2,6-Diaminopurine (aka 2-Aminoadenine, Base D, DAP) replaced.
- As was found in 2016, is 2' Desoxyarchaeosin (dG +) in the genome of several bacteria and Escherichia phage 9g (ICTV: Escherichia virus 9g , genus Nonagvirus , family Siphoviridae ) present.
- 6-methylisoxanthopterin
- 5-hydroxyuracil
Natural modified bases (methylations, etc.)
Modified bases are also found in natural DNA. In particular, methylations of canonical bases are examined in the context of epigenetics :
- First, in 1925, 5-methylcytosine (m 5 C) was found in the genome of Mycobacterium tuberculosis . In the genome of the Xanthomonas oryzae bacteriophage Xp12 (Xanthomonas phage XP-12, Fam Siphoviridae ) and the Halovirus ΦH ( Halobacterium virus phiH , genus Myohalovirus , Myoviridae ), the entire cystosine contingent has been replaced by 5-methylcytosine.
- A complete replacement of cytosine by 5- glycosylhydroxymethylcytosine ( syn. Glycosyl-5-hydroxymethylcytosine) in phages T2, T4 and T6 of the species Escherichia virus T4 (genus T4virus , subfamily Tevenvirinae of the family Myoviridae ) was observed in 1953.
- As was discovered in 1955, is N 6 -methyladenine (6mA, m 6 A) in the DNA of E. coli bacteria present.
- N 6 -carbamoylmethyladenine was described in 1975 in the bacteriophages Mu (ICTV: species Escherichia virus Mu , formerly Enterobacteria phage Mu ; genus Muvirus , obsolete Mulikevirus in the family Myoviridae ) and Lambda-Mu.
- 7-methylguanine (m 7 G) was described in 1976 in phage DDVI (' Enterobacteria phage DdVI ' alias ' DdV1 ', genus T4virus ) of Shigella disenteriae .
- N 4 -Methylcytosine (m 4 C) in DNA was described in 1983 (in Bacillus centrosporus ).
- In 1985 5-hydroxycytosine was found in the genome of the Rhizobium phage RL38JI.
- α-Putrescinylthymine (Alpha-Putrescinylthymine, putT) and α-Glutamylthymidine (Alpha-Glutamylthymidine) occur in the genome of both the Delftia phage ΦW-14 (Phi W-14, species ' Dellftia virus PhiW14 ', genus ' Ionavirus ', family Myovrirdae ) as well as the Bacillus phage SP10 (also family Myoviridae ).
- 5-Dihydroxypentyluracil was found in the Bacillus phage SP15 (also SP-15, family Myoviridae ).
The function of these non-canonical bases in DNA is not known. They work at least in part as a molecular immune system and help protect bacteria from infection by viruses.
Non-standard and modified bases in microbes are not everything:
- Four modifications of the cytosine residues in human DNA have also been reported. These modifications consist of the addition of the following groups:
- Methyl (-CH 3 )
- Hydroxymethyl (-CH 2 OH)
- Formyl (-CHO)
-
Carboxyl (–COOH)
It is assumed that these modifications have regulatory functions, keyword epigenetics .
- Uracil is found in the centromere regions of at least two human chromosomes (6 and 11).
Synthetic bases
In the laboratory, DNA (and also RNA) was provided with additional artificial bases. The aim is mostly so unnatural base pairs ( English unnatural pairs base , UBP) to produce:
- In 2004, DNA was generated that instead of the four standard nucleobases (A, T, G and C) contained an extended alphabet with six nucleobases (A, T, G, C, dP and dZ). With these two new bases dP stands for 2-amino -8- (1'-β- D -2'-deoxyribofuranosyl) - imidazo [1,2- a ] - 1,3,5-triazine - 4 (8 H ) -one and dZ for 6-amino-5-nitro-3- (1'-β- D -2'-deoxyribofuranosyl) -2 (1 H ) -pyridone.
- In 2006, a DNA with bases extended by a benzene group or a naphthyl group was examined for the first time (either xDNA or xxDNA or yDNA or yyDNA, depending on the position of the extension groups).
- Yorke Zhang et al. reported at the turn of the year 2016/2017 on semi-synthetic organisms with a DNA that was expanded to include the bases X (alias NaM) and Y '(alias TPT3) or the (deoxyribo) nucleotides dX (dNaM) and dY' (dTPT3), who mate with each other. This was preceded by experiments with pairings based on the bases X and Y (alias 5SICS), i. H. of nucleotides dX and dY (alias d5SICS). Other bases that can pair with 5SICS are FEMO and MMO2.
- At the beginning of 2019, there were reports on DNA and RNA with eight bases each (four natural and four synthetic), which are all assigned to each other in pairs ( Hachimoji DNA ).
Enantiomers
DNA occurs in living things as D -DNA; L DNA as enantiomer ( Spiegelmer ) can be synthesized, however, (the same applies analogously for RNA). L -DNA is broken down more slowly by enzymes than the natural form, which makes it interesting for pharmaceutical research .
Genetic information content and transcription

DNA molecules play an important role as information carriers and “docking points” for enzymes that are responsible for transcription . Furthermore, the information of certain DNA segments, as it is present in operative units such as the operon, is important for regulatory processes within the cell.
Certain sections of DNA, the so-called genes , encode genetic information that influences the structure and organization of the organism. Genes contain “blueprints” for proteins or molecules that are involved in protein synthesis or the regulation of the metabolism of a cell. The sequence of the bases determines the genetic information. This base sequence can be determined by means of sequencing, for example using the Sanger method.
The base sequence (base sequence) of a gene segment of the DNA is initially overwritten by the transcription into the complementary base sequence of a so-called ribonucleic acid molecule (abbreviated to RNA). In contrast to DNA, RNA contains the sugar ribose instead of deoxyribose and the base uracil instead of thymine, but the information content is the same. So-called mRNAs are used for protein synthesis , single-stranded RNA molecules that are transported from the cell nucleus into the cytoplasm , where protein synthesis takes place ( see protein biosynthesis ).
According to the so-called “one-gene-one-protein hypothesis”, the sequence of a protein molecule is read from a coding section on the DNA. However, there are regions of DNA that each encode several proteins by using different reading frames during transcription. In addition, various isoforms of a protein can be produced by alternative splicing (subsequent cutting of the mRNA).
In addition to the coding DNA (genes), there is non-coding DNA , which makes up over 90 percent of the total DNA of a cell in humans.
The storage capacity of the DNA has not yet been technically reproduced. With the information density in a teaspoon of dried DNA, the current world population could be reconstructed about 350 times.
DNA replication

According to the so-called semiconservative principle, the DNA can duplicate (replicate) itself with the help of enzymes . The double-stranded helix is separated by the enzyme helicase after it has been de- coiled by the topoisomerase . The resulting single strands serve as a template (template) for the complementary complementary strand to be synthesized, which attaches to them.
DNA synthesis, i.e. H. the binding of the nucleotides to be linked is carried out by enzymes from the group of DNA polymerases . A nucleotide to be linked must be present in the triphosphate compound - i.e. as deoxyribonucleoside triphosphate. By splitting off two phosphate parts, the energy required for the binding process is released.
The enzyme helicase forms a replication fork, two diverging single strands of DNA. In its area, an RNA primer , which is synthesized by the primase enzyme, marks the starting point of the new DNA synthesis. The DNA polymerase successively attaches nucleotides to this RNA molecule that are complementary to those of the DNA single strands.
The linkage of the new nucleotides to a complementary single strand of DNA can only run in the 5 '→ 3' direction on the two old strands and therefore does so without interruption along the old 3 '→ 5' strand in the direction of the increasingly opening Replication fork.
The synthesis of the new strand on the old 5 '→ 3' strand, on the other hand, cannot take place continuously towards the replication fork, but only away from it, also in the 5 '→ 3' direction. At the beginning of the replication, the old double strand is only open a little, so that only a short piece of new complementary DNA can ever develop on the second strand - in the “unsuitable” opposite direction.
Since a DNA polymerase only links about 1000 nucleotides in each case, it is necessary to synthesize the entire complementary strand in individual pieces. When the replication fork has opened a little wider, a new RNA primer is therefore attached again directly to the fork on the second single strand and initiates the next DNA polymerase.
When synthesizing the 3 '→ 5' strand, a new RNA primer is therefore required for each DNA synthesis unit. The primer and the associated synthesis unit are known as the Okazaki fragment . The RNA primers required for the start of replication are then enzymatically degraded. This creates gaps in the new DNA strand, which are filled with DNA nucleotides by special DNA polymerases.
Finally, the enzyme ligase links the not yet connected new DNA segments into a single strand.
Mutations and other DNA damage
Mutations of DNA segments - for example exchange of bases for others or changes in the base sequence - lead to changes in the genetic make-up , some of which can be fatal for the organism concerned.
In rare cases, however, such mutations are also beneficial; they then form the starting point for the change in living beings in the course of evolution . By means of recombination during sexual reproduction , this change in DNA even becomes a decisive factor in evolution: the eukaryotic cell usually has several sets of chromosomes, i.e. i.e. a double strand of DNA is present at least twice. The mutual exchange of parts of these DNA strands, the crossing-over in meiosis , can create new properties.
DNA molecules can be damaged by various influences. Ionizing radiation , such as UV or γ radiation , alkylation and oxidation can chemically change the DNA bases or lead to strand breakage. These chemical changes may affect the pairing properties of the affected bases. Many of the mutations during replication come about in this way.
Some common DNA damage are:
- the formation of uracil from cytosine with the spontaneous loss of an amino group through hydrolysis : like thymine, uracil is complementary to adenine.
- Thymine-thymine dimer damage caused by the photochemical reaction of two successive thymine bases in the DNA strand through UV radiation , for example from sunlight . This damage is likely a major cause of skin cancer development .
- the formation of 8-oxoguanine through oxidation of guanine: 8-oxoguanine is complementary to both cytosine and adenine. During replication , both bases can be incorporated opposite 8-oxoguanine.
Due to their mutagenic properties and their frequent occurrence (estimates amount to 10 4 to 10 6 new damage per cell and day), DNA damage must be removed from the genome in good time . For this, cells have an efficient DNA repair system. It eliminates damage using the following strategies:
- Direct damage reversal: An enzyme reverses the chemical change in the DNA base.
- Base excision repair: The faulty base, for example 8-oxoguanine, is cut out of the genome. The resulting free space is re-synthesized using the information in the opposite strand.
- Nucleotide excision repair: A larger strand containing the damage is cut out of the genome. This is re-synthesized using the information in the opposite strand.
- Homologous recombination : If both DNA strands are damaged, the genetic information from the second chromosome of the homologous chromosome pair is used for the repair.
- Replication with special polymerases: DNA polymerase η can, for example, replicate error-free via TT dimer damage. People in whom polymerase η does not work or works only to a limited extent often suffer from xeroderma pigmentosum , a hereditary disease that leads to extreme sensitivity to sunlight.
Denaturation
The base pairing of DNA is denatured in various cellular processes . The base pairing is broken in sections by various DNA-binding proteins , e.g. B. in replication or transcription . The location of the start of denaturation is known as the denaturation bubble and is described in the Poland-Scheraga model . However, the DNA sequence , stiffness and torsion are not included. The lifespan of a denaturation bubble is between one microsecond and one millisecond.
In the laboratory, DNA can be denatured by physical and chemical methods. DNA is denatured by formamide , dimethylformamide , guanidinium salts, sodium salicylate , sulfoxide , dimethyl sulfoxide (DMSO), various alcohols , propylene glycol and urea , usually in combination with heat. Concentrated solutions of sodium hydroxide also denature DNA. With the chemical methods, the melting temperature of the double-stranded DNA is lowered.
DNA purification and detection
DNA can be purified by DNA purification , e.g. B. by DNA extraction , are separated from other biomolecules . The qualitative detection of DNA (which DNA is present) is mostly done by a polymerase chain reaction , an isothermal DNA amplification , a DNA sequencing , a Southern blot or an in situ hybridization . The quantitative detection (how much DNA is present) is mostly done by qPCR ; in the case of purified samples with only one DNA sequence, a concentration can also be measured by photometry at a wavelength of 260 nm. An extinction of 1 of a purified DNA solution corresponds to a concentration of 50 µg / mL for double- stranded DNA, 33 µg / mL for single-stranded DNA and lower for single-stranded oligonucleotides , depending on the composition of the nucleobases (see DNA extraction #Quantification ). DNA can be stained by intercalating dyes such as ethidium bromide , propidium iodide or SYBR Green I and by furrow-binding dyes such as DAPI , pentamidine , lexitropsine , netropsin , distamycin , Hoechst 33342 or Hoechst 33258 . Less specifically bound DNA dyes and staining methods are e.g. B. methylene blue , the carbocyanine dye Stains-All or the silver dye . By chromosome combing the DNA may be stretched and aligned.
"Old" DNA
Remnants of genome molecules in dead organisms are referred to as aDNA (“ancient DNA”) if there are no more direct relatives of the organism being sampled. Human DNA is also referred to as aDNA if the individual died at least 75 years before the sample was examined.
See also
- Xenonucleic acid , XNA, plus:
- LNA
- Peptide nucleic acid (PNA)
- Morpholino
- Dideoxyribonucleoside triphosphates (ddNTPs): Artificial intermediates in DNA sequencing according to Sanger
- Deoxyadenosine mono-arsenate (dAMAs) see GFAJ-1 §Discussion about the incorporation of arsenic in biomolecules (questionable incorporation in DNA in Halomonas species GFAJ-1, see also Halomonas titanicae )
literature
- Chris R. Calladine and others: DNA - the molecule and how it works. 3. Edition. Spektrum Akademischer Verlag, Heidelberg 2005, ISBN 3-8274-1605-1 .
- Ernst Peter Fischer: In the beginning there was the double helix. James D. Watson and the New Science of Life. Ullstein, Berlin 2004, ISBN 3-548-36673-2 .
- Ernst Peter Fischer: The Genome. An introduction. Fischer, Frankfurt am Main 2002, ISBN 3-596-15362-X .
- James D. Watson: The Double Helix . Rowohlt, Reinbek 1997, ISBN 3-499-60255-5 .
- James D. Watson: Gene, Girls, and Gamow. Memories of a genius. Piper, Munich 2003, ISBN 3-492-04428-X .
- James D. Watson: In the beginning there was the double helix . Ullstein, Berlin 2003, ISBN 3-550-07566-9 .
- James D. Watson, M. Gilman, J. Witkowski, M. Zoller: Recombined DNA. 2nd Edition. Spektrum Akademischer Verlag, Heidelberg 1993, ISBN 3-86025-072-8 .
- Tomas Lindahl : Instability and decay of the primary structure of DNA. In: Nature . Volume 362, 1993, pp. 709-715. doi: 10.1038 / 362709a0 .
- W. Wayt Gibbs: Precious items in the DNA scrap. In: Spectrum of Science . No. 2, 2004, pp. 68-75. (on-line)
- W. Wayt Gibbs: DNA isn't everything. In: Spectrum of Science. No. 3, 2004, pp. 68-75. (on-line)
- Hans-Jürgen Quadbeck-Seeger: The structure of DNA - a model project. In: Chemistry in Our Time . Volume 42, 2008, pp. 292-294. doi: 10.1002 / ciuz.200890046 .
Web links
- Video: DNA isolation from tomatoes
- DNA Interactive - Cold Spring Harbor Institute and Howard Hughes Medical Institute site (an excellent introduction to the subject)
- DNA from the Beginning of the Dolan DNA Learning Center
- "DNA from the Beginning" (German)
- “Translator” to find the coded amino acid to the coding base triplet or vice versa
- “Translator” of an entire DNA segment into the coded amino acids
- Harvard cracks DNA storage, crams 700 terabytes of data into a single gram
Notes and individual references
- ↑ In linguistic usage, deoxyribonucleic acid is predominantly referred to using the English abbreviation for deoxyribonucleic acid ; the German abbreviation DNS is called "obsolete" by the Duden . See Duden. The German spelling. Volume 1. 26th edition. Mannheim 2013.
- ↑ Bärbel Häcker: DNS. In: Werner E. Gerabek , Bernhard D. Haage, Gundolf Keil , Wolfgang Wegner (eds.): Enzyklopädie Medizingeschichte. De Gruyter, Berlin / New York 2005, ISBN 3-11-015714-4 , p. 316 f .; here: p. 316.
- ↑ Hubert Mania: A victim of the scientific prejudices of his time. The DNA was discovered as early as 1869 in the Tübingen Renaissance castle. On: Telepolis. April 17, 2004.
- ^ Richard Altmann : Ueber nucleic acids. In: Archives for Anatomy and Physiology. Physiological department. Leipzig 1889, pp. 524-536.
- ^ P. Levene: The structure of yeast nucleic acid . In: J Biol Chem . tape 40 , no. 2 , 1919, p. 415-424 ( jbc.org ).
- ↑ Joachim Gerlach: Hartwig Kuhlenbeck † In: Würzburger medical history reports. Volume 2, 1984, pp. 269-273, here: p. 271.
- ↑ W. Astbury: Nucleic acid . In: Symp. SOC. Exp. Bbl . tape 1 , no. 66 , 1947.
- ^ O. Avery, C. MacLeod, M. McCarty: Studies on the chemical nature of the substance inducing transformation of pneumococcal types. Inductions of transformation by a deoxyribonucleic acid fraction isolated from pneumococcus type III . In: J Exp Med . tape 79 , no. 2 , 1944, pp. 137–158 , doi : 10.1084 / jem.149.2.297 .
- ↑ A. Hershey, M. Chase: Independent functions of viral protein and nucleic acid in growth of bacteriophage . In: J Gen Physiol . tape 36 , no. 1 , 1952, pp. 39-56 , PMID 12981234 ( rupress.org [PDF]).
- ↑ 50 years of the double helix . Spectrum of Science , February 28, 2003
- ↑ February 28: The Day Scientists Discovered the Double Helix . Scientific American , February 28, 2013.
- ↑ a b J. D. Watson, FH Crick: Molecular structure of nucleic acids. A structure for deoxyribose nucleic acid. In: Nature . Volume 171, No. 4356, 1953, pp. 737-738. PMID 13054692 , full text (PDF; 368 kB)
- ↑ Katharina Kramer: On the trail of life. In: GEO compact. No. 7, 2006.
- ^ Information from the Nobel Foundation on the award ceremony in 1962 .
- ↑ www.ensembl.org, Homo sapiens : Database status from February 2009. (website in English).
- ^ R. Wing, H. Drew, T. Takano, C. Broka, S. Tanaka, K. Itakura, R. Dickerson: Crystal structure analysis of a complete turn of B-DNA . In: Nature . tape 287 , no. 5784 , 1980, pp. 755-758 , PMID 7432492 .
- ↑ C. Pabo, R. Sauer: Protein-DNA recognition . In: Annu Rev Biochem . tape 53 , p. 293-321 , PMID 6236744 .
- ↑ SC Ha, K. Lowenhaupt, A. Rich, YG Kim, Kim KK: Crystal structure of a junction between B-DNA and Z-DNA Reveals two extruded bases . In: Nature . tape 437 , 2005, pp. 1183-1186 , PMID 16237447 .
- ↑ a b Peter Yakovchuk, Ekaterina Protozanova, Maxim D. Frank-Kamenetskii: Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. In: Nucleic Acids Research . Volume 34, No. 2, 2006, pp. 564-574. doi: 10.1093 / nar / gkj454 PMID 16449200
- ↑ Gerhard Steger (Ed.): Bioinformatics: Methods for the prediction of RNA and protein structures. Birkhäuser Verlag, Basel, Boston, Berlin 2003.
- ↑ S Kiljunen, K Hakala, E Pinta, S Huttunen, P Pluta, A Gador, H Lönnberg, M Skurnik: Yersiniophage phiR1-37 is a tailed bacteriophage having a 270 kb DNA genome with thymidine replaced by deoxyuridine . In: Microbiology . 151, No. Pt 12, December 2005, pp. 4093-4102. doi : 10.1099 / mic.0.28265-0 . PMID 16339954 .
- ↑ J Uchiyama, I Takemura-Uchiyama, Y Sakaguchi, K Gamoh, S Kato, M Daibata, T Ujihara, N Misawa, S Matsuzaki: Intragenus generalized transduction in Staphylococcus spp. by a novel giant phage . In: The ISME Journal . 8, No. 9, September 2014, pp. 1949–1952. doi : 10.1038 / ismej.2014.29 . PMID 24599069 . PMC 4139722 (free full text).
- ↑ Taxonomy - Bacillus phage PBS2 (Bacteriophage PBS2) (SPECIES) , on UniProt, accessed February 21, 2019.
- ↑ Yersinia phage phiR1-RT , on: Virus-Host DB.
- ↑ Molnár P, Marton L, Izrael R, Pálinkás HL, Vértessy BG (2018) Uracil moieties in Plasmodium falciparum genomic DNA. FEBS Open Bio 8 (11): 1763-1772.
- ↑ Andrew MQ King et al. (Ed.): Virus Taxonomy: Classification and Nomenclature of Viruses (PDF) Ninth Report of the International Committee on Taxonomy of Viruses
- ↑ a b c ICTV : dsDNA Viruses> Myoviridae , in: ICTV 9th Report (2011)
- ↑ a b E Casella, O Markewych, M Dosmar, W Heman: Production and expression of dTMP-enriched DNA of bacteriophage SP15 . In: J Virology , 28 (3), 1978, pp. 753-766, PMID 153409 , PMC 525799 (free full text), ResearchGate .
- ↑ David H. Roscoe: Synthesis of DNA in phage-infected Bacillus subtilis , in: Virology 38 (4), September 1969, pp. 527-537, doi: 10.1016 / 0042-6822 (69) 90173-1 .
- ↑ Bacillus phage SP-15 , on: Virus Host DB
- ↑ MS Walker, M. Mandel: PMC 353962 (free full text) Biosynthesis of 5- (4'5'-dihydroxypentyl) uracil as a nucleoside triphosphate in bacteriophage SP15-infected Bacillus subtilis. In: J Virol. 1978 Feb; 25 (2): pp. 500-509. PMC 353962 (free full text), PMID 146749 .
- ↑ J. Marmurm et al .: Unique Properties of Nucleic Acid from Bacillus subtilis Phage SP-15 . In: Nature New Biology , 1972, 239, p. 68. doi: 10.1038 / newbio239068a0 - SP-15 infects B. subtilis and B. licheniformis .
- ↑ a b c Andrew M. Kropinski et al .: The Sequence of Two Bacteriophages with Hypermodified Bases Reveals Novel Phage-Host Interactions . In: Viruses , May 2018, 10 (5), p. 217. PMC 5977210 (free full text), PMID 29695085 , PMC 5977210 (free full text).
- ↑ L Simpson: A base called J . In: Proceedings of the National Academy of Sciences of the United States of America . 95, No. 5, March 1998, pp. 2037-2038. bibcode : 1998PNAS ... 95.2037S . doi : 10.1073 / pnas.95.5.2037 . PMID 9482833 . PMC 33841 (free full text).
- ^ P Borst, R Sabatini: Base J: discovery, biosynthesis, and possible functions . In: Annual Review of Microbiology . 62, 2008, pp. 235-251. doi : 10.1146 / annurev.micro.62.081307.162750 . PMID 18729733 .
- Jump up ↑ M Cross, R Kieft, R Sabatini, M Wilm, M de Kort, GA van der Marel, JH van Boom, F van Leeuwen, P Borst: The modified base J is the target for a novel DNA-binding protein in kinetoplastid protozoans . In: The EMBO Journal . 18, No. 22, November 1999, pp. 6573-6581. doi : 10.1093 / emboj / 18.22.6573 . PMID 10562569 . PMC 1171720 (free full text).
- ↑ C DiPaolo, R Kieft, M Cross, R Sabatini: Regulation of trypanosome DNA glycosylation by a SWI2 / SNF2-like protein . In: Molecular Cell . 17, No. 3, February 2005, pp. 441-451. doi : 10.1016 / j.molcel.2004.12.022 . PMID 15694344 .
- ↑ S Vainio, PA Genest, B ter Riet, H van Luenen, P Borst: Evidence did J-binding protein 2 is a thymidine hydroxylase catalyzing the first step in the biosynthesis of DNA base J . In: Molecular and Biochemical Parasitology . 164, No. 2, April 2009, pp. 157-161. doi : 10.1016 / j.molbiopara.2008.12.001 . PMID 19114062 .
- ↑ LM Iyer, M Tahiliani, A Rao, L Aravind: Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids . In: Cell Cycle . 8, No. 11, June 2009, pp. 1698-1710. doi : 10.4161 / cc.8.11.8580 . PMID 19411852 . PMC 2995806 (free full text).
- ↑ HG van Luenen, C Farris, S Jan, PA Genest, P Tripathi, A Velds, RM Kerkhoven, M Nieuwland, A Haydock, G Ramasamy, S Vainio, T Heidebrecht, A Perrakis, L Pagie, B van Steensel, PJ Myler , P Borst: Glucosylated hydroxymethyluracil, DNA base J, prevents transcriptional readthrough in Leishmania . In: Cell . 150, No. 5, August 2012, pp. 909-921. doi : 10.1016 / j.cell.2012.07.030 . PMID 22939620 . PMC 3684241 (free full text).
- ↑ DZ Hazel Baker, S Buratowski: Transcription: base J blocks the way . In: Current Biology . 22, No. 22, November 2012, p. R960-2. doi : 10.1016 / j.cub.2012.10.010 . PMID 23174300 . PMC 3648658 (free full text).
- ↑ US20060270005A1 , Genomic library of cyanophage s-21 and functional analysis
- ↑ Han Xia et al .: Freshwater cyanophages . In: Virologica Sinica , 28 (5), pp. 253-259, October 2013, doi: 10.1007 / s12250-013-3370-1
- ^ MHV van Regenmortel et al. : ICTV 7th Report (PDF) 2000
- ↑ RS Safferman, RE Cannon, PR Desjardins, BV Gromov, R. Haselkorn, LA Sherman, M. Shilo: Classification and Nomenclature of Viruses of Cyanobacteria . In: Intervirology . 19, No. 2, 1983, pp. 61-66. doi : 10.1159 / 000149339 . PMID 6408019 .
- ↑ Roger Hull, Fred Brown, Chris Payne (Eds.): Virology: A Directory and Dictionary of Animal, Bacterial and Plant Viruses, Macmillan Reference Books, ISBN 978-1-349-07947-6 , doi: 10.1007 / 978-1 -349-07945-2 , hit
- ↑ IY Khudyakov, MD Kirnos, NI Alexandrushkina, BF Vanyushin: Cyanophage S-2L contains DNA with 2,6-diaminopurine substituted for adenine . In: Virology . 88, No. 1, 1978, pp. 8-18. PMID 676082 .
- ↑ JJ Thiaville, SM Kellner, Y Yuan, G Hutinet, PC Thiaville, W Jumpathong, S Mohapatra, C Brochier-Armanet, AV Letarov, R Hillebrand, CK Malik, CJ Rizzo, PC Dedon, V de Crécy-Lagard: Novel genomic island modifies DNA with 7-deazaguanine derivatives . In: Proceedings of the National Academy of Sciences of the United States of America . 113, No. 11, 2016, pp. E1452-1459. bibcode : 2016PNAS..113E1452T . doi : 10.1073 / pnas.1518570113 . PMID 26929322 . PMC 4801273 (free full text).
- ^ TB Johnson, RD Coghill: Pyrimidines. CIII. The discovery of 5-methylcytosine in tuberculinic acid, the nucleic acid of the tubercle bacillus. . In: Journal of the American Chemical Society . 47, 1925, pp. 2838-2844.
- ↑ Pieter-Jan Ceyssens: The Genome and Structural Proteome of YuA, a New Pseudomonas aeruginosa phage Resembling M6. In: Journal of Bacteriology . 190 (4): pp. 1429-1435, March 2008, doi: 10.1128 / JB.01441-07 .
- ↑ TT Kuo, TC Huang, MH Teng: 5-Methylcytosine replacing cytosine in the deoxyribonucleic acid of a bacteriophage for Xanthomonas oryzae . In: Journal of Molecular Biology . 34, No. 2, 1968, pp. 373-375. PMID 5760463 .
- ↑ Heike Vogelsang-Wenke, Dieter Oesterhelt: Isolation of a halobacterial phage with a fully cytosine-methylated genome . In: MGG Molecular & General Genetics . 211, No. 3, March 1988, pp. 407-414. doi : 10.1007 / BF00425693 .
- ^ GR Wyatt, SS Cohen: The bases of the nucleic acids of some bacterial and animal viruses: the occurrence of 5-hydroxymethylcytosine . In: The Biochemical Journal . 55, No. 5, 1953, pp. 774-782. PMID 13115372 . PMC 1269533 (free full text).
- ^ DB Dunn, JD Smith: Occurrence of a new base in the deoxyribonucleic acid of a strain of Bacterium coli . In: Nature . 175, No. 4451, 1955, pp. 336-337. PMID 13235889 .
- ↑ E Bremer et al .: Lambda placMu: a transposable derivative of bacteriophage lambda for creating lacZ protein fusions in a single step . In: J Bacteriol. , 1984 Jun, 158 (3), pp. 1084-1093, PMC 215554 (free full text), PMID 6327627
- ↑ Diego de Mendoza et al .: Cloning of mini-Mu bacteriophage in cosmids: in vivo packaging into phage lambda heads . In: Gene , Vol. 39, Issue 1, 1985, pp. 55-59
- ^ B Allet, AI Bukhari: Analysis of bacteriophage mu and lambda-mu hybrid DNAs by specific endonucleases . In: Journal of Molecular Biology . 92, No. 4, 1975, pp. 529-540. PMID 1097703 .
- ^ II Nikolskaya, NG Lopatina, SS Debov: Methylated guanine derivative as a minor base in the DNA of phage DDVI Shigella disenteriae . In: Biochimica et Biophysica Acta . 435, No. 2, 1976, pp. 206-210. PMID 779843 .
- ↑ A Janulaitis, S Klimasauskas, M Petrusyte, V Butkus: Cytosine in DNA modification by Bcn I methylase yields N4-methylcytosine . In: FEBS Letters . 161, No. 1, 1983, pp. 131-134. PMID 6884523 .
- ↑ D Swinton, S Hattman, R Benzinger, V Buchanan-Wollaston, J Beringer: Replacement of the deoxycytidine residues in Rhizobium bacteriophage RL38JI DNA . In: FEBS Letters . 184, No. 2, 1985, pp. 294-298. PMID 2987032 .
- ↑ Ionavirus , ICTV Proposals
- ↑ KL Maltman, J Neuhard, RA Warren: 5 - [(hydroxymethyl) -O-pyrophosphoryl] uracil, to intermediate in the biosynthesis of alpha-putrescinylthymine in deoxyribonucleic acid of bacteriophage phi W-14 . In: Biochemistry . 20, No. 12, 1981, pp. 3586-3591. PMID 7260058 .
- ↑ YJ Lee, N Dai, SE Walsh, S Müller, ME Fraser, KM Kauffman, C Guan, IR Corrêa Jr, PR Weigele: Identification and biosynthesis of thymidine hypermodifications in the genomic DNA of widespread bacterial viruses . In: Proc Natl Acad Sci USA, April 3, 2018, 115 (14), pp. E3116-E3125. doi: 10.1073 / pnas.1714812115 , PMID 29555775 .
- ↑ H. Hayashi, K. Nakanishi, C. Brandon, J. Marmur: Structure and synthesis of dihydroxypentyluracil from bacteriophage SP-15 deoxyribonucleic acid . In: J. Am. Chem. Soc. , 1973, 95, pp. 8749-8757. doi: 10.1021 / ja00807a041 .
- ^ T Carell, MQ Kurz, M Müller, M Rossa, F Spada: Non-canonical bases in the genome: The regulatory information layer in DNA . In: Angewandte Chemie (International Ed. In English) . 2017. doi : 10.1002 / anie.201708228 . PMID 28941008 .
- ↑ X Shu, M Liu, Z Lu, C Zhu, H Meng, S Huang, X Zhang, C Yi: Genome-wide mapping reveals that deoxyuridine is enriched in the human centromeric DNA . In: Nat Chem Biol , 2018, doi: 10.1038 / s41589-018-0065-9 .
- ↑ Kyung Hyun Lee, Kiyofumi Hamashima, Michiko Kimoto, Ichiro Hirao: Genetic alphabet expansion biotechnology by creating unnatural base pairs . In: Current Opinion in Biotechnology , Volume 51, June 2018, pp. 8-15, ScienceDirect , ResearchGate , PMID 29049900 , doi: 10.1016 / j.copbio
- ↑ AM Sismour, S. Lutz, JH Park, MJ Lutz, PL Boyer, SH Hughes, SA Benner: PCR amplification of DNA containing non-standard base pairs by variants of reverse transcriptase from Human Immunodeficiency Virus-1 . In: Nucleic acids research . Volume 32, number 2, 2004, pp. 728-735, doi: 10.1093 / nar / gkh241 , PMID 14757837 , PMC 373358 (free full text).
- ↑ Z. Yang, D. Hutter, P. Sheng, AM Sismour and SA Benner: Artificially expanded genetic information system: a new base pair with an alternative hydrogen bonding pattern. () Nucleic Acids Res. , 34, 2006, pp. 6095-6101. PMC 1635279 (free full text)
-
↑ Z. Yang, AM Sismour, P. Sheng, NL Puskar, SA Benner: Enzymatic incorporation of a third nucleobase pair . In: Nucleic acids research , Volume 35, Number 13, 2007, pp. 4238-4249,
doi: 10.1093 / nar / gkm395 , PMID 17576683 , PMC 1934989 (free full text). -
^ SR Lynch, H. Liu, J. Gao, ET Kool: Toward a designed, functioning genetic system with expanded-size base pairs: solution structure of the eight-base xDNA double helix . In: Journal of the American Chemical Society , Volume 128, No. 45, November 2006, pp. 14704-14711,
doi: 10.1021 / ja065606n . PMID 17090058 . PMC 2519095 (free full text). - ↑ Yorke Zhang, Brian M. Lamb, Aaron W. Feldman, Anne Xiaozhou Zhou, Thomas Lavergne, Lingjun Li, Floyd E. Romesberg: A semisynthetic organism engineered for the stable expansion of the genetic alphabet . In: PNAS , 114 (6), February 7, 2017, pp. 1317-1322; first published January 23, 2017, doi: 10.1073 / pnas.1616443114 , Ed .: Clyde A. Hutchison III, The J. Craig Venter Institute
- ↑ Researchers breed "Frankenstein" microbes . scinexx, January 24, 2017
- ↑ Indu Negi, Preetleen Kathuria, Purshotam Sharma, Stacey D. Wetmore: How do hydrophobic nucleobases differentiate from natural DNA nucleobases? Comparison of structural features and duplex properties from QM calculations and MD simulations . In: Phys. Chem. Chem. Phys. , 2017, 19, pp. 16305-16374, doi: 10.1039 / C7CP02576A
- ↑ Shuichi Hoshika, Nicole A. Leal, Myong-Jung Kim, Myong-Sang Kim, Nilesh B. Karalkar, Hyo-Joong Kim, Alison M. Bates, Norman E. Watkins Jr., Holly A. Santa Lucia, Adam J. Meyer , Saurja DasGupta, Joseph A. Piccirilli, Andrew D. Ellington, John SantaLucia Jr., Millie M. Georgiadis, Steven A. Benner: Hachimoji DNA and RNA: A genetic system with eight building blocks. In: Science , 363 (6429), February 22, 2019, pp. 884-887, doi: 10.1126 / science.aat0971 .
- ↑ Daniela Albat: DNA with eight letters . scinexx. Extended Code of Life . Wissenschaft.de (bdw online), both from February 22, 2019.
- ↑ W. Purschke, F. Radtke, F. Klein Young, S. Klussmann: A DNA Spiegelmer to staphylococcal enterotoxin B . In: Nucleic Acids Research. Volume 31, No. 12, 2003, pp. 3027-3032. doi: 10.1093 / nar / gkg413 , PMID 12799428
- ↑ Gosuke Hayashi, Masaki Hagihara, Kazuhiko Nakatani: Application of L-DNA as a molecular tag . In: Nucleic Acids Symposium Series , Vol. 49, No. 1, 2005, pp. 261-262. doi: 10.1093 / wet / 49.1.261 , PMID 17150733
- ↑ Leonard M. Adelmann: Calculating with the DNA. In: Spectrum of Science. November 1998, p. 77, (full text)
- ↑ François Sicard, Nicolas Destainville, Manoel Manghi: DNA denaturation bubbles: Free-energy landscape and nucleation / closure rates . In: The Journal of Chemical Physics . 142, No. 3, January 21, 2015, p. 034903. arxiv : 1405.3867 . doi : 10.1063 / 1.4905668 .
- ^ Simon Lieu: The Poland-Scheraga Model. (2015): 0-5. Massachusetts Institute of Technology, May 14, 2015.
- ^ C. Richard, AJ Guttmann: Poland – Scheraga Models and the DNA Denaturation Transition. In: Journal of Statistical Physics. 115, 2004, p. 925, doi: 10.1023 / B: JOSS.0000022370.48118.8b .
- ↑ Grégoire Altan-Bonnet, Albert Libchaber, Oleg Krichevsky: Bubble Dynamics in Double-Stranded DNA . In: Physical Review Letters . 90, No. 13, April 1, 2003. doi : 10.1103 / physrevlett.90.138101 .
- ↑ J. Marmur, PO Ts'O: Denaturation of deoxyribonucleic acid by formamide. In: Biochimica et Biophysica Acta . Volume 51, July 1961, pp. 32-36, PMID 13767022 .
- ↑ a b c d Prog Nucleic Acid Res & Molecular Bio . Academic Press, 1963, ISBN 978-0-08-086289-7 , p. 267.
- ↑ a b Hyone-Myong Eun: Enzymology Primer for Recombinant DNA Technology. Elsevier, 1996, ISBN 978-0-08-053113-7 , p. 67.
- ↑ Wolfgang Hennig: Genetics. Springer-Verlag, 2013, ISBN 978-3-662-07430-5 ( limited preview in Google book search).