Myoviridae
| Myoviridae | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
||||||||||||||
| Systematics | ||||||||||||||
|
||||||||||||||
| Taxonomic characteristics | ||||||||||||||
|
||||||||||||||
| Scientific name | ||||||||||||||
| Myoviridae | ||||||||||||||
| Left | ||||||||||||||
|

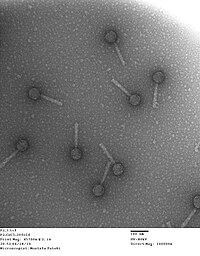
The Myoviridae family comprises six genera of bacteriophage with a molecule of double-stranded DNA as a genome . The members of this family have an icosahedral capsid and a contractile tail part; Because of this contractile property, the family name derives from the Greek myos (μυός) for muscle . Within the Caudovirales order , the Myoviridae are characterized by particularly large, sometimes elongated capsids (e.g. T4 phage : 111 × 78 nm) and a very large genome (34-169 kbp ). The most important representative is the T4 phage ( Enterobacteria Phage T4 ), which was discovered early on, and whose special morphology and genetic properties played a prominent role in molecular biology research. The T4 DNA ligase derived from this phage is still used today in molecular biology.
Myoviridae have besides bacteria and archaea to host .
Systematics
The following system according to ICTV (status 2018b.v2 from March 2019, updated with status 2019.v1 from March 2020) only includes part of the associated species:
Family Myoviridae
-
- Genus Firehammervirus (obsolete Cp220virus , Cp220likevirus , Cp220-like viruses )
- Species Campylobacter virus CP220
- Genus Cp8virus (alias Cp8unalikevirus )
- Species Campylobacter virus CP81
-
- Genus Kolesnikvirus (obsolete Ea214virus )
- Genus Felixounavirus (obsolete Felixo1virus , Felixounalikevirus )
- Species Salmonella virus FelixO1
- Genus mooglevirus
- Species Citrobacter virus Moogle
- Genus Suspvirus
-
- Genus Hpunavirus (outdated Hp1virus , Hpunalikevirus , Hp1likevirus )
- Species Haemophilus phage HP1
- Genus Peduovirus (obsolete P2virus , P2likevirus , P2-like viruses , 13 species)
- Species Escherichia virus P2 (alias Enterobacteria Phage P2 , P2-Phage )
-
- Genus Pecentum virus (obsolete P100 virus )
- Genus Okubovirus (obsolete Spo1virus , Spounalikevirus , SPO1-like viruses , two species)
- Species Bacillus virus SPO1 (alias Bacillus phage SPO1 )
- Genus tsarbombavirus
-
- Genus Karamvirus (obsolete Cc31virus )
- Genus Jiaodavirus (obsolete Jd18virus )
- Genus Dhakavirus (obsolete Js98virus )
- Genus Slopekvirus (obsolete Kp15virus )
- Genus moon virus
- Genus Krischvirus (obsolete Rb49virus )
- Genus Mosigvirus (obsolete Rb69virus )
- Genus Gelderland virus (outdated S16 virus )
- Genus Schizotequatrovirus (obsolete Schizot4virus )
- Genus Gaprivervirus (obsolete Sp18virus )
- Genus Tequatrovirus (obsolete T4virus , T4likevirus , T4-like viruses , 13 species)
- Species Escherichia virus T4 (alias Enterobacteria Phage T4 , T4 phage , with 12 subtypes, including T2 phage )
-
- Genus Certrevirus (outdated Cr3virus )
- Genus Seunavirus (outdated Se1virus )
- Species Salmonella virus PVPSE1 (alias Salmonella phage PVP-SE1 )
- Genus Vequintavirus (outdated V5virus )
-
- Genus Abouovirus
- Genus Acionnavirus
- Genus Agricanvirus (obsolete Agrican357virus )
- Genus Ahtivirus
- Genus Alcyoneus virus
- Genus Alexandravirus
- Genus Anamdong virus
- Genus Anaposvirus
- Genus Aokuang virus
- Genus Aphroditevirus
- Genus Asterius virus
- Genus atlauavirus
- Genus Aurunvirus
- Genus Baikal virus
- Genus Barbavirus
- Genus Bcepmuvirus (alias Bcepmulikevirus , Bcepmu-like viruses )
- Species Burkholderia virus BcepMu
- Genus Bellamy virus
- Genus bendigovirus
- Genus biquartavirus
- Genus Bixzunavirus (obsolete Bxzunalikevirus , Bxz1virus , I3likevirus , I3-like viruses , I3-like viruses )
-
- Species Mycobacterium virus Bxz1 (alias Mycobacterium-Phage Bxz1 ) (*)
- Mycobacterium phage Cali
- Mycobacterium phage Catera
- Mycobacterium phage Rizal
- Mycobacterium phage ScottMcG
- Mycobacterium phage spud
- Species Mycobacterium virus I3 (alias Mycobacterium phage I3 )
- Species " Mycobacterium phage Myrna " (proposed)
- Genus Brigitvirus
- Genus Brizovirus
- Genus Brunovirus
- Genus Busan virus
- Genus carpasinavirus
- Genus Chakrabartyvirus
- Genus Charybdisvirus
- Genus Chiangmaivirus (outdated Rsl2virus )
- Genus Emdodecavirus (obsolete M12virus )
- Genus Cymopoleiavirus
- Genus Derbicus virus
- Genus Elvirus (obsolete Ellikevirus )
- Species Pseudomonas virus EL (scientific Pseudomonas virus EL , obsolete Pseudomonas phage EL ) (*)
- Genus Emdodecavirus
- Genus Eneladus virus
- Genus Eponavirus
- Genus Erskinevirus (obsolete Eah2virus )
- Genus Eurybiavirus
- Genus Ficleduovirus
- Genus Fluff Dravirus
- Genus Gofduovirus
- Genus Goslar virus
- Genus Hapunavirus (outdated Hapunalikevirus , Hap1likevirus )
- Species Halomonas virus HAP1
- Genus Heilongjiang virus
- Genus Iapetus virus
- Genus iodovirus
- Species Iodobacter virus PLPE (obsolete Iodobacter phage phiPLPE ) (*)
- Genus Ionavirus
- Genus Jedunavirus
- Genus Jilinvirus (outdated Cvm10virus )
- Genus Jimmer virus
- Genus Kanaloavirus
- Genus Kleczkowskavirus (outdated Rheph4virus )
- Genus lagaffe virus
- Genus Leucotheavirus
- Genus Liban virus
- Genus Llyrvirus
- Genus Loughborough virus
- Genus Lubbockvirus (outdated Cd119virus , Phicd119likevirus )
- Species Clostridium virus phiCD119
- Genus machinavirus
- Genus Marthavirus
- Genus Mazu virus
- Genus metrivirus
- Genus Louseafarmvirus (obsolete Rslunavirus )
- Genus mimas virus
- Genus Mushuvirus
- Genus Muvirus (outdated Mulikevirus , Mu-like viruses , Mu-like phages , Mu-like viruses , 2 species)
- Species Escherichia virus Mu (alias Enterobacteria-Phage Mu )
- Genus Myohalovirus (outdated Phihlikevirus , phiH-like viruses , PhiH-like viruses )
- Species Halobacterium virus phiH (alias Halobacterium phage PhiH , phage ϕH - also ΦH , spelling: phage øH )
- Species " Halobacterium phage Hs1 " (provisional)
- Genus Naesvirus
- Species Burkholderia virus Bcep1
- Species Burkholderia virus Bcep43
- Species Burkholderia virus Bcep781 (*, formerly genus Vidavervirus ; obsolete Bcep78virus , Bcep78likevirus , Bcep78-like viruses )
- Species Xanthomonas virus OP2
- Genus Namakavirus
- Genus Nankokuvirus (outdated Kpp10virus )
- Genus Neptune virus
- Genus Nereus virus
- Species Synechococcus virus ACG2014bSyn9311C4
- Genus Nerrivikvirus
- Genus nodal virus
- Species Synechococcus virus SPM2 (alias Synechococcus phage S-PM2 , Bacteriophage S-PM2 ) (*)
- Genus Noxifervirus
- Genus Obolenskvirus (outdated Ap22virus )
- Genus Otagovirus
- Genus Pakpunavirus
- Genus Palaemonvirus
- Genus Pbunavirus (obsolete Pbunalikevirus , PB1likevirus )
- Species Pseudomonas virus PB1 (alias Pseudomonas phage PB1 )
- Genus peat virus
- Genus Petsuvirus
- Genus Phabquatrovirus
- Genus Phapecoctavirus
- Genus Phikzvirus (outdated Phikzlikevirus , PhiKZ-like viruses , PhiKZ-like viruses )
- Species Pseudomonas virus phiKZ (alias Pseudomonas phage PhiKZ , Bacteriophage ϕKZ - also ΦKZ , φKZ )
- Species Pseudomonas virus PA7
- Species Pseudomonas virus SL2
- Species " Pseudomonas Phage 201phi2-1 " (alias " Pseudomonas chlororaphis phage 201phi2-1 ")
- Genus plaisance virus
- Genus Polybotosvirus
- Genus Pontus virus
- Genus Popoffvirus (obsolete Plpelikevirus , Dwarf myoviruses )
- Species Aeromonas virus 56 (obsolete Aeromonas phage 56 )
- Genus Punavirus (obsolete P1virus , Punalikevirus , P1likevirus , P1-like viruses , 2 species)
- Species Escherichia virus P1 (alias Enterobacteria phage P1 , P1 phage )
- Species Aeromonas virus 43 (alias Aeromonas phage 43 )
- Genus Qingdaovirus
- Genus Radnorvirus (outdated Arv1virus )
- Genus Ripduovirus
- Genus Risingsunvirus
- Genus Rosemount virus
- Genus Saclayvirus
- Genus Salacisavirus
-
- Species Prochlorococcus virus PSSM2 (alias Prochlorococcus phage P-SSM2 , Cyanophage P-SSM2 )
- P-SSM2 Fd (Fd like ferredoxin , infects Prochlorococcus marinus )
- Genus Salmond Virus
- Genus Sasquatch virus
- Genus Schmittlotzvirus
- Genus Seoulvirus (outdated Spn3virus )
- Species Salmonella virus SPN3US (obsolete Salmonella phage SPN3US )
- Genus Shalavirus
- Genus Svunavirus
- Genus Tabernarius virus
- Genus Tamkungvirus
- Genus Taranis virus
- Genus Tefnutvirus
- Genus Tegunavirus (outdated Tg1virus )
- Genus thaumas virus
- Genus thetis virus
- Species Synechococcus virus SSM1 (alias Synechococcus phage S-SM1 , Cyanophage S-SM1 )
- Genus thorn virus
- Genus Tidunavirus
- Genus Tijeunavirus
- Genus Toutatis virus
- Genus Tulanevirus (obsolete Secunda5virus )
- Genus vellamovirus
- Genus Vhmlvirus
- Genus Viunavirus
- Genus Wellington virus
- Genus Wifcevirus
- Genus Winkler virus
- Genus Yokohamavirus (obsolete Msw3virus )
- Genus Yoloswagvirus
- Gender not determined:
- Species Bacillus virus G
- Species Bacillus virus PBS1
- Species Microcystis virus Ma-LMM01
-
- Genus " Cbasmlikevirus "
- Species " Cellulophaga phage phiSM " (*)
- Species " Cellulophaga phage phi3: 1 "
- Species " Cellulophaga phage phi3ST: 2 "
- Species " Cellulophaga phage phi38: 2 "
- Species " Cellulophaga phage phi47: 1 "
- Genus " Cyanomyoviruses " (informal)
- Species " Synechococcus phage Syn9 " (alias " Bacteriophage Syn9 ", " Cyanophage Syn9 ")
- Species " Prochlorococcus phage P-SSM4 " (alias " Cyanophage P-SSM4 ")
- Genus " Hfunalikevirus "
- Species " Halovirus HF1 " (*)
- Species " Halovirus HF2 "
- Genus " Rv5likevirus "
- Species " Enterobacteria phage phi92 "
- Species " Escherichia phage rV5 " (*)
- Species " Escherichia phage vB_EcoM_FV3 "
- Genus " Sfv and relatives " (informal, to be distinguished from the species Rabbit fibroma virus alias Shope fibroma virus , genus Leporipoxvirus , Poxviridae )
- Species " Enterobacteria phage SfV " (alias " Shigella phage SfV ", " Shigella flexneri bacteriophage V ") (*)
- Species " Escherichia phage P27 "
- without proposed genus:
- Species " Aggregatibacter phage Aaphi23 " (alias " Bacteriophage Aaphi23 ", suggested)
- Species " Bdellovibrio phage phi1402 "
- Species " Bdellovibrio phage phi1422 "
- Species " Pectobacterium phage ZF40 "
- Species " Pseudomonas phage OBP " (alias Pseudomonas fluorescens bacteriophage OBP , suggested)
- Species " Sphingomonas Phage PAU "
- Species " Vibrio phage 138 "
- Species " Vibrio phage CP-T1 "
- Species " Yersinia phage PY100 "
-
- " Kabirphage "
- " Mahaphage " with the subgroup
- " Lak-Phages " with the representatives Lak-A1, Lak-A2, Lak-B1 to Lak-B9 and Lak-C1
- " Biggiephage "
- " Dakhmphage "
- " Kyodaiphage "
- " Kaempephage "
- " Jabbarphage "
- " Enormephage "
- " Judaphage "
- " Whopperphage "
Displacements:
- Genus Vi1virus (obsolete Viunalikevirus ) with species Bacteriophage ϕMAM1 was moved to the new Caudovirales family Ackermannviridae , subfamily Cvivirinae .
- Genus Siminovitchvirus (outdated Cp51virus ) was moved to the new Caudovirales family Herelleviridae , subfamily Spounavirinae .
- The genera Agatevirus , Bequatrovirus (obsolete B4virus ), Bastillevirus , Caeruleovirus (obsolete Bc431virus ), Nitunavirus (obsolete Nit1virus ) with species Bacillus virus NIT1 and Wphvirus have also been moved to the new family Herelleviridae , but subfamily Bastillevirinae .
- The new subfamily Twortvirinae of the Herelleviridae now contains the genera Baoshanvirus , Harbinvirus , Kayvirus , Sciuriunavirus , Sepunavirus (out of date Sep1virus ), Silviavirus , Twortvirus (alias Twortlikevirus , Twort-like viruses ) with the type species Staphylococcus virus Twort
literature
- AMQ King, MJ Adams, EB Carstens, EJ Lefkowitz (Eds.): Virus Taxonomy. Ninth Report of the International Committee on Taxonomy of Viruses. Amsterdam 2012, ISBN 978-0-12-384684-6 , pp. 46-62.
- CM Fauquet, MA Mayo et al .: Eighth Report of the International Committee on Taxonomy of Viruses. London / San Diego 2004.
Web links
- Myoviridae: Genera and Species (NCBI)
- Myoviridae Family Picture Gallery (Big Picture Book of Viruses)
Individual evidence
- ↑ a b c d ICTV: ICTV Master Species List 2019.v1 , New MSL including all taxa updates since the 2018b release, March 2020 (MSL # 35)
- ↑ a b c d ICTV: ICTV Taxonomy history: Enterobacteria phage T4 , EC 51, Berlin, Germany, July 2019; Email ratification March 2020 (MSL # 35)
- ↑ NCBI: Synechococcus phage S-PM2 (species)
- ↑ Pseudomonas phage 201phi2-1 , on: Virus-Host DB, TAX: 198110
- ↑ Proteomes - Pseudomonas phage 201phi2-1 , on: UniProt
- ↑ NCBI: Pseudomonas phage 201phi2-1 (species)
- ↑ NCBI: Prochlorococcus virus PSSM2 (species)
- ↑ a b Denise Brehm: Viruses Use Bacteria for Reproduction , on: SciTechDaily from January 26, 2012
- ↑ a b Qinglu Zeng, Sallie W. Chisholm: Marine Viruses Exploit Their Host's Two-Component Regulatory System in Response to Resource Limitation , in: Curr Biol Volume 22, No. & nbs; 2, CellPress of January 24, 2012, pp. 124-128, doi: 10.1016 / j.cub.2011.11.055 , PDF , Supplement 1
- ↑ Ian J. Campbell, Jose Luis Olmos Jr., Weijun Xu, Dimithree Kahanda, Joshua T. Atkinson, Othneil Noble Sparks, Mitchell D. Miller, George N. Phillips Jr., George N. Bennett, Jonathan J. Silberg: Prochlorococcus phage ferredoxin: Structural characterization and electron transfer to cyanobacterial sulfite reductases - Phage Fd characterization and host SIR interactions, in: J. Biol. Chem., ASBMB Publications from May 19, 2020, doi: 10.1074 / jbc.RA120.013501 , PDF
- ↑ Beneath the Ocean's Surface, a Virus Is Hijacking the Most Abundant Organism on Earth , on: SciTechDaily from June 6, 2020, source: Rice University
- ↑ NCBI: Synechococcus virus S-SM1 (species)
- ↑ S. B. Santos, A. M. Kropinski, P.-J. Ceyssens, H.-W. Ackermann, A. Villegas, R. Lavigne, V. N. Krylov, CM Carvalho, E. C. Ferreira, J. Azeredo: Genomic and Proteomic Characterization of the Broad-Host-Range Salmonella Phage PVP-SE1: Creation of a New Phage Genus . In: Journal of Virology . 85, No. 21, 2011, pp. 11265-11273. doi : 10.1128 / JVI.01769-10 . PMID 21865376 . PMC 3194984 (free full text).
- ↑ L. Truncaite, E. Šimoliūnas, A. Zajančkauskaite, L. Kaliniene, R. Mankevičiūte, J. Staniulis, V. Klausa, R. Meškys: Bacteriophage vB_EcoM_FV3: A new member of "rV5-like viruses" . In: Archives of Virology . 157, No. 12, 2012, pp. 2431-2435. doi : 10.1007 / s00705-012-1449-x . PMID 22907825 .
- ↑ A. Cornelissen, S. C. Hardies, O. V. Shaburova, V. N. Krylov, W. Mattheus, A. M. Kropinski, R. Lavigne: Complete Genome Sequence of the Giant Virus OBP and Comparative Genome Analysis of the Diverse KZ-Related Phages . In: Journal of Virology . 86, No. 3, 2011, pp. 1844-1852. doi : 10.1128 / JVI.06330-11 . PMID 22130535 . PMC 3264338 (free full text).
- ↑ CM Mizuno, F. Rodriguez-Valera, N. E. Kimes, R. Ghai: Expanding the Marine Virosphere Using Metagenomics . In: PLoS Genetics . 9, No. 12, 2013, p. E1003987. doi : 10.1371 / journal.pgen.1003987 . PMID 24348267 . PMC 3861242 (free full text).
- ↑ K. Holmfeldt, N. Solonenko, M. Shah, K. Corrier, L. Riemann, NC Verberkmoes, M. B. Sullivan: Twelve previously unknown phage genera are ubiquitous in global oceans . In: Proceedings of the National Academy of Sciences . 110, No. 31, 2013, pp. 12798-12803. doi : 10.1073 / pnas.1305956110 . PMID 23858439 . PMC 3732932 (free full text).
- ↑ NCBI: unclassified Myoviridae
- ↑ NCBI: Synechococcus phage syn9 (species)
- ↑ NCBI: Cyanophage Syn9 (species)
- ↑ NCBI: Prochlorococcus phage P-SSM4 (species)
- ↑ NCBI: Rabbit fibroma virus (species)
- ↑ NCBI: Enterobacteria phage SfV (species)
- ↑ NVBI: Sphingomonas phage PAU (species)
- Jump up ↑ Richard Allen White III, Curtis A. Suttle: The Draft Genome Sequence of Sphingomonas paucimobilis Strain HER1398 (Proteobacteria), Host to the Giant PAU Phage, Indicates That It Is a Member of the Genus Sphingobacterium (Bacteroidetes) , in: Genome Announc. 1 (4), July-August 2013, e00598-13, doi: 10.1128 / genomeA.00598-13 , PMID 23929486 , PMC 3738902 (free full text)
- ↑ Basem Al-Shayeb, Rohan Sachdeva, L. Chen, Jillian F. Banfield et al. : Clades of huge phages from across Earth's ecosystems , in: Nature from February 12, 2020, doi: 10.1038 / s41586-020-2007-4
- ↑ Ed Yong: A Huge Discovery in the World of Viruses , on: The Atlantic, February 20, 2020
- ↑ Audra E. Devoto, Joanne M. Santini et al. : Megaphages infect Prevotella and variants are widespread in gut microbiomes , in: Nature Microbiology, Volume 4, pp. 693-700, January 28, 2019, doi: 10.1038 / s41564-018-0338-9 , especially Table 1 and Supplementary Figure 11