Protein synthesis

Protein biosynthesis (PBS) is the formation of new proteins in cells and thus the process of gene expression that is central to all living beings , in which proteins are built up from amino acids according to genetic information .
The actual synthesis of a protein from its building blocks, the proteinogenic amino acids, takes place on the ribosomes and is also referred to as translation , since the base sequence of a messenger RNA (mRNA) is translated into the sequence of amino acids of a peptide . This is done by continuously assigning the anticodon of a transfer RNA (tRNA) to one codon of the mRNA and binding its individually transported amino acid to the neighboring one ( peptide bond ), so that a chain with a characteristicAmino acid sequence arises. This polypeptide can unfold in the surrounding medium to form a structure of structured three-dimensional form , the native protein . Often it is changed afterwards, post-translationally modified through splits, alterations and additions .
While in prokaryotic cells ( procytes ) the circular DNA is free in the cytosol and the ribosomal protein synthesis usually takes place immediately and promptly on the mRNA template that has just been transcribed, the situation in eukaryotic cells ( eucytes ) is more complicated. For the genome, which is distributed over several chromosomes , the cell nucleus (nucleus) has its own compartment , in the caryoplasm of which transcription also takes place. The primarily extracted RNA copy (hnRNA) is first stabilized, revised and prepared for nuclear export before it passes through a nuclear pore as mRNA and reaches the cytoplasm , which contains the subunits of the ribosomes. This spatial division and the multi-step process path thus allow additional ways of modifying an (hn) RNA post-transcriptionally and regulating gene expression or excluding certain RNA templates from protein synthesis ( gene shutdown ).
Some types of bacteria , archaea and fungi can build special proteins than ribosomal protein synthesis multi-enzyme complexes a nonribosomal peptide synthesis allow (NRPS).
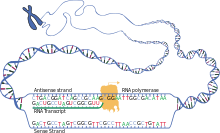
transcription
In the first step for protein biosynthesis in a cell, sections of genes on the double-stranded DNA are searched for, read and transcribed into single-stranded RNA molecules. During this process, the sequence of nucleobases of DNA ( adenine , guanine , cytosine , thymine ) is complementarily assigned the nucleobases of RNA building blocks ( uracil , cytosine, guanine, adenine). Ribose instead of deoxyribose and uracil instead of thymine appear in the RNA transcript that is then linked to form a strand . The genetic information is contained in the base sequence, a codogen on the DNA is transcribed to a codon on the messenger ribonucleic acid, called mRNA for short .
For transcription of a gene in addition to several other factors is an RNA polymerase necessary as enzyme which the continuous structure of the RNA polymer depends on the DNA template catalyzed . The base-pairing ribonucleoside triphosphates ( UTP , CTP , GTP and ATP ) assigned to the template as building blocks are linked to one another to form a polynucleotide of an RNA, each with splitting off two phosphate groups of the triphosphates . There can be different types of RNA polymerase for the transcription of genes that code for proteins by means of an mRNA, and for those of other genes, for example for the formation of an rRNA or a tRNA .
In eukaryotes , transcription takes place in the nucleus ; therefore, the mRNA has to be exported from the nucleus into the cytosol , since translation is carried out there. Prokaryotes, on the other hand, do not have a core compartment, as transcription takes place here in addition to translation in the cell plasma.
Post-transcriptional modification
- Splice
- In the case of eukaryotes, after the pure transcription, the non-coding introns contained in the pre-mRNA thus created must be cut out so that only the required exons remain. This process is called splicing . Consensus sequences are used to identify the introns. During splicing, different snRNPs bind in the area of the introns and exon-intron junctions. With the formation of the spliceosome, these lead to the cleavage of the phosphodiester bonds and thus to the cutting out of the introns. At the same time the exons are ligated. Splicing also occurs with rRNA and tRNA.
- Capping
- During this time, so-called capping takes place, during which the stability of the RNA is increased. A so-called 5'-cap structure is attached, with the 5 'end of the pre-mRNA being synthesized being converted to a structure called a "cap" and the mRNA before digestion by 5'-exonucleases and Phosphatases protects.
- Polyadenylation
- During polyadenylation , the poly (A) tails are attached to the newly created 3 'end of the RNA (up to 250 nucleotides long). This poly (A) tail facilitates the export of the mRNA into the cytoplasm and also protects the 3 'end from enzymatic degradation.
- RNA edition
- With the RNA edition , individual or several nucleobases of the RNA molecule are changed (modified), inserted (inserted) or cut out (deleted) after transcription. For example, editing can result in a new stop codon on the mRNA that is upstream of the previous one; the translation then breaks off here and the shorter isoform of a protein is formed. RNA editing only occurs in some organisms, cells or cell organelles and is often limited to particular nucleotide sequences.
Translation or protein synthesis
Under Translation refers to the translation of the base sequence of the mRNA in the amino acid sequence of the protein to the ribosome happens. In the mRNA, three consecutive bases, a base triplet , form a codon within the open reading frame that codes for an amino acid (see genetic code ). On the ribosome, the codons are translated into amino acids according to their sequence and these are linked sequentially to form a polypeptide .
To form a peptide bond between two amino acids, they have to be brought into close proximity to one another. This requires the surface of a large supramolecular structure. This task is fulfilled by the ribosomes, composed of a small and a large subunit, which forms two adjacent binding sites: the A-site and the P-site.
Since there is no structural relationship between a codon and the associated amino acid, an intermediate piece is required which, on the one hand, binds the amino acid and, on the other hand, recognizes the associated codon on the mRNA. This mediating function is performed by transfer ribonucleic acid molecules, various tRNAs , as amino acid “transporters” with recognition regions. They have two exposed binding sites, the amino acid binding site and the anticodon . The amino acid binding sites of the tRNAs are specifically loaded with the appropriate amino acid by the aminoacyl-tRNA synthetases . The tRNA recognizes the complementary codon on the mRNA with the anticodon and specifically binds to it.
The translation process as such can be divided into three phases: the initial phase, the elongation phase and finally the termination:
- Initial phase
- If a previously synthesized mRNA reaches a ribosome , the small subunit of the ribosome migrates along the mRNA until it encounters the start codon AUG. The corresponding methionine tRNA with the anticodon UAC is attached to the codon ( initiation complex ).
- Elongation phase
- With the cleavage of GTP , the large subunit of the ribosome now also accumulates and elongation begins.
- During the initiation phase, the methionine tRNA is on the P binding site, so that the next tRNA can accumulate in the A binding site. A peptidyl transferase links the methionine of the first tRNA with the amino acid of the subsequent tRNA; this formation of a dipeptide takes place in the A-binding site. Eventually the ribosome units migrate one base triplet.
- The tRNA with the dipeptide is now on the P-binding site from which it has displaced the very first, now unloaded tRNA, and the next tRNA, whose anticodon is complementary to the base triplet of the mRNA strand, can again attach to the free A-binding site fits.
- Termination
- If a ribosome moving along the mRNA encounters one of the three stop codons , translation initially comes to a standstill because there are no matching tRNA molecules which are encoded for an amino acid (suppression). They are replaced by so-called termination or release factors (RFs), which bind to the A-site and change the substrate specificity of the peptidyl transferase in such a way that a water molecule is activated instead of an AA tRNA. Its nucleophilic attack on the bond between the peptide chain and tRNA finally releases the synthesized protein and separates the mRNA from the ribosome.
Co- and post-translational modification
The polypeptide chains of some proteins are already changed during translation (cotranslational) by special enzymes, but in most cases proteins are only modified after translation has been completed (post-translational). While chaperones influence the shaping process of protein folding, on which the association to protein complexes also depends, a cell also has a multitude of possibilities to specifically change the structure of a protein, to create functionally different protein species and thus to expand the proteome through modifications .
These modifications include the cleavage of individual terminal amino acids or the longer peptide sequences in the case of precursor proteins , the introduction of additional bonds, e.g. B. disulfide bridges between cysteine residues , or functional groups, such as hydroxylations of amino acids ( proline to 4- hydroxyproline by the prolyl-4-hydroxylase , lysine to hydroxylysine by the lysyl hydroxylase ), as well as oxidations (e.g. covalent cross-links by means of lysine residues by the lysyl oxidase ), Carboxylation or decarboxylation and numerous others. For example, glycoproteins are formed through glycosylation , lipoproteins through acylation and prenylation .
The individual steps of modification are each catalyzed by special enzymes, the occurrence of which is often limited to certain organelles, cells or tissues. In addition, the sequence of modifying steps or their temporal course can be varied, depending on the cell environment, development phase or environmental conditions. The collagen molecule, for example, goes through a series of post-translational modifications, some of which only take place in the extracellular space.
Protein targeting and protein transport
Because many proteins as the destination ( english target ) is not the cytosol, but the extracellular space , the cell membrane , the organelles such as chloroplasts , mitochondria , peroxisomes , nucleus or endoplasmic reticulum have the cell has several mechanisms to spend the proteins there. These proteins usually contain an N- or C-terminal signal sequence , which can be structured very differently depending on the target mechanism. In some cases there is no terminal signal sequence, but rather internal signals of the peptide chain, which determine the target location of the protein.
- Proteins that target the endoplasmic reticulum (ER) carry a specific N-terminal sequence that is recognized by a protein-RNA complex, the Signal Recognition Particle (SRP). The SRP-peptide-ribosome complex is then recruited to the endoplasmic reticulum where it is recognized and bound. Translation continues through the membrane. The attached ribosomes give the impression of a “rough ER”. See cotranslational protein transport . The quality control of the newly synthesized protein takes place in the endoplasmic reticulum .
- Proteins that need to be brought into the chloroplasts have an N-terminal signal sequence that is usually early phosphorylated . The proteins Hsp70 , 14-3-3 and Toc64 can also play a role in recognition and transmission through interaction with the protein precursor. After arriving on the surface of the chloroplast, the protein-precursor complex is recognized by receptor structures of the translocon apparatus of the outer chloroplast membrane ( Translocon Of Outer Chloroplast Membrane , TOC). Under GTP - hydrolysis of the protein is then imported into the intermembrane space, or directly by the Translokonapparat ( TIC ) of the inner chloroplast membrane into the stroma imported. For the import into the membrane or the lumen of the thylakoids at least 4 paths are used, which are called Sec-dependent , SRP-dependent , delta-pH / Tat-dependent or spontaneous .
- So far, three different import routes have been described for the mitochondrion for yeast and animal cells:
- The pre-sequence import route, whose proteins carry an N -terminal amphiphilic alpha helix. These proteins are mostly intended for the matrix, the inner membrane or the intermembrane space.
- The carrier protein import route for proteins in the inner membrane that carry various internal signals.
- The import route of the proteins of the outer envelope membrane, which is used to integrate proteins with a beta barrel motif. Here, too, there are sequence-internal signals.
- All three import routes start at the mitochondrial translocation apparatus in the outer membrane ( TOM ), which has different receptors. The Tom20 and Tom22 receptors recognize the N-terminal signal and pass the precursor protein on to the Tom40 pore . The Tom70 receptor recognizes the internal signals of the proteins that are intended for the outer membrane.
- After the import into the intermembrane space, the paths separate: The proteins with the beta-barrel motif, which are intended for the outer membrane, are integrated into the membrane by the SAM complex (sorting and assembly machinery). The proteins of the other two import routes are directed to different TIM complexes: proteins with a presequence are recognized by the TIM23 complex, whereas proteins for the inner membrane are recognized by the TIM22 complex .
- The pre-sequence is by the enzyme MPP ( English mitochondrial processing peptidase away).
In addition to the signal sequences described above, glycosylation enables targeting for incorporation into the cell membrane or for exocytosis . Both routes usually lead via Golgi vesicles .
See also
Web links
Individual evidence
- ↑ Matthias Strieker, Alan Tanović, Mohamed A. Marahiel: nonribosomal peptide synthetases: structures and dynamics. In: Current opinion in structural biology. Volume 20, number 2, April 2010, pp. 234-240, doi : 10.1016 / j.sbi.2010.01.009 , PMID 20153164 .
- ^ Gavin J. Williams: Engineering polyketide synthases and nonribosomal peptide synthetases. In: Current opinion in structural biology. Volume 23, number 4, August 2013, pp. 603-612, doi : 10.1016 / j.sbi.2013.06.012 , PMID 23838175 .
- ↑ a b c Translation . ( Spektrum.de [accessed June 15, 2018]).