tRNA
tRNA is short for transfer RNA . Transfer RNAs are short ribonucleic acids (RNA) . The length of mature tRNAs is usually between 73 and 95 nucleotides . During translation , they use the base triplet of their anticodon to convey the correct amino acid to the corresponding codon on the mRNA .
structure
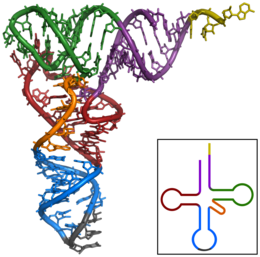
tRNA molecules usually consist of 73 to 95 nucleotides of a single RNA strand. In addition to the four basic building blocks of RNA ( adenosine , uridine , cytidine and guanosine ), tRNA contains a number of different, modified standard bases . For example, one has the nucleosides dihydrouridine (D), inosine (I), 2-thiouridine (s 2 U), 4-thiouridine (s 4 U), pseudouridine (Ψ), N 4 -acetylcytidine (ac 4 C) and ribothymidine (T) identified. The bases are often also methylated to different degrees (cf. DNA methylation ).
Pairings of conjugating nucleobases occur in every tRNA molecule ; they form double-stranded regions. In a two-dimensional representation, tRNA has a cloverleaf-like secondary structure . This structure forms a base and three loops: a dihydrouracil - a TΨC- and anticodon loop (see picture on the right.).
- D-arm (dihydrouracil arm): The so-called dihydrouracil loop at the end of the D arm owes its name to the dihydrouracil residues that it often contains. It is usually shown on the left in figures. However, dihydrouracil residues are not always present in every dihydrouracil loop. The Dihydrouracil loop is primarily used to recognize the tRNA by the aminoacyl-tRNA synthetase.
- Low anticodon: On the anticodon loop there is a specific base triplet , the so-called anticodon . The loop always consists of five conjugating pairs and seven unpaired bases, three of which form the anticodon. This interacts with the complementary codon of the mRNA to be read (see section Function).
- T loop (TΨC loop): The TΨC loop shown in the figures on the right contains seven unpaired bases, three of which form the eponymous sequence TΨC. T stands for thymidine, Ψ for pseudouridine and C for cytidine.
- Variable loop: The so-called variable loop is located between the TΨC arm and the anticodon arm. This varies in length depending on the tRNA.
- Acceptor Strain: The acceptor strain is phosphorylated at its 5 'end. Its 3 'end always shows the sequence CCA-3'. The 3 'end, an adenosine, presents its ribose, which has an important physiological role. At this end, the 3'-OH group of ribose, the carboxy group of a corresponding amino acid is esterified and thus bound to the tRNA (see section Function).
The modified bases are most frequently found among the non-conjugated bases of the trefoil structure. The actual three-dimensional tertiary structure resembles an “L” (upside down in the figure), whose two arms form the amino acid acceptor stem and the anticodon loop. The functionally important sections are therefore as far apart as possible. The three-dimensional structure of tRNA from yeast was elucidated in 1974 with a resolution of 300 pm (3 Å ) independently of two groups led by Rich and Klug. In 2000 a structure with an improved resolution of 1.93 Å was published.
A tRNA Ala from S. cerevisiae
function
During translation in protein biosynthesis , the appropriate amino acid must be attached to the peptide chain in the ribosome according to the genetic code for each base triplet on the mRNA. This task is mediated by the tRNA. There is at least one, but often several different tRNAs for each amino acid.
Loading of the tRNA with an amino acid
With ATP consumption, tRNAs are specifically loaded with the associated amino acid at the 3 'end by the respective aminoacyl-tRNA synthetase, depending on their sequence . For this purpose, the carboxy group of the amino acid is attached in an ester bond to the 3'- hydroxyl group of the ribose of the adenosine, so that an aminoacyl group is formed. Often the aminoacyl-tRNA synthetase recognizes the anticodon on the tRNA. However, other structural elements can also play a role in the recognition, mainly the acceptor stem.
A special case are the tRNAs that are loaded with alanine (tRNA Ala ). Regardless of the organism, the tRNA Ala has a G – U base pair at positions 3 and 70 in the acceptor stem. If one of these two bases is exchanged for another by (targeted) mutagenesis, the resulting tRNA can no longer be loaded with alanine by the alanyl-tRNA Ala synthetase.
Initially it was assumed that in all organisms there is exactly one aminoacyl-tRNA synthetase for each amino acid, which can load all tRNAs belonging to the corresponding amino acid. However, it was later discovered that some organisms lack one or more aminoacyl-tRNA synthetases, but that they can still incorporate the corresponding amino acids into their proteins and also have the necessary tRNAs. These must therefore have a different mechanism for loading these tRNAs. Many archaea, bacteria, chloroplasts and mitochondria lack an aminoacyl-tRNA synthetase for glutamine . Instead, the aminoacyl-tRNA synthetase for glutamate binds it to both tRNA for glutamate and tRNA for glutamine. The “incorrectly” bound glutamate to the tRNA for glutamine is then converted into glutamine by a transamidase (Glu-tRNA Gln amidotransferase).
Incorporation of amino acids into the nascent chain
The aminoacylated tRNAs are used by the ribosomes for protein biosynthesis. If the anticodon matches the corresponding base codon of the messenger RNA (mRNA) , the tRNA can attach itself there and the transported amino acid can attach to the resulting protein .
According to the genetic code , a tRNA would have to exist for every base triplet that codes an amino acid and is not a stop codon - generally 61. However, it was found that the number of tRNAs deviates significantly downwards. The exact number differs in the organisms, but it is no less than 31 and no more than 41. Nevertheless, in all organisms all base triplets are used for protein coding. This deviation is explained by the wobble theory .
| Base of the anticodon (tRNA) | Base of the codon (mRNA) |
|---|---|
| C. | G |
| A. | U |
| G | U or C |
| I. | U, C or A |
| U | A or G; in chloroplasts and mitochondria also U or C |
This theory, which goes back to Francis Crick , states that the first two bases of the codon and the last two bases of the anticodon form hydrogen bonds according to the "classic" Watson-Crick base pairing; G always pairs with C or A with U. According to Crick's investigations, the possibility of base pairing between the third base of the codon and the first base of the anticodon can be extended and does not follow this strict rule. One also speaks of the "wobble position".
For example, if the first base of the anticodon is a U, it can recognize an A or G of the codon. Sometimes this first base is also an inosine; it can interact with both U, C and A of the codon. The results are summarized in the table on the right.
The first base of the anticodon is often modified. This affects the recognition of the corresponding third base of the codon. For example, if the first base of the anticodon is a lysidine (k 2 C), this is only recognized by an A in bacteria. In the mitochondria of some eukaryotes, base pairs 5-formylcytidine (f 5 C) with A or G. In echinoderms and the mitochondria of cuttlefish it has been discovered that 7-methylguanosine (m 7 G) can interact with all four standard bases.
Accordingly, due to the fact that the genetic code is a degenerate code in which several base triplets can code for the same amino acid, often only two bases are necessary for recognition, since base triplets for the same amino acid often only differ in one base.
Initiator tRNAs
In the synthesis of a polypeptide, the starting process ( initiation ) places different demands on the tRNA than the lengthening ( prolongation ) of the chain. Since the synthesis usually starts at a codon AUG (coding for methionine), the cell requires a special tRNA suitable for initiation for the start codon AUG. This tRNA i , also known as initiator tRNA, differs from another methionine-specific tRNA Met , which is necessary for the recognition of the AUG codon within a reading frame. Although both tRNAs are loaded with methionine by the same methionyl-tRNA synthetase, tRNA Met cannot be used for the start codon, only tRNA i Met .
In bacteria, for example E. coli , the initiator tRNA is given as tRNA i fMet . The "f" in the name indicates that the tRNA loaded with methionine is modified with a formyl residue . This reaction is catalyzed by a methionyl-tRNA-formyltransferase ( EC 2.1.2.9 ), during which N 10 -formyl tetrahydrofolate (10-CHO-THF) is converted to tetrahydrofolic acid (THF). The tRNA i fMet differs in important points from the tRNA Met , so that the latter is not recognized by the formyltransferase and therefore only tRNA i fMet is used for the initiation. So there is no base pairing between the first cytosine and the adenine in the acceptor strain (see picture). There are three consecutive G – C base pairs in the anticodon strain. Finally, the initiator tRNA in the D loop has a CCU sequence. Together, these differences create a slightly different conformation.
The initiator tRNA in eukaryotes, as well as in the archaea, is not formylated. After translation, the protein formed can be modified. In bacteria, the N-terminal formyl residue is usually split off . The methionine at the N-terminus of the polypeptide can then also be split off, as can eukaryotes and archaea.
Biosynthesis of tRNA
tRNA genes
Organisms vary in the number of their tRNA genes on their genome. The nematode Caenorhabditis elegans , a typical model organism for genetic studies, has 29,647 genes in its nucleus genome, 620 of which code for tRNA. The baker's yeast has 275 tRNA genes in their genome .
In prokaryotes , several genes are usually grouped together in operons . The genes can code for protein sequences or various RNA products, including tRNA. As a rule, several tRNAs are combined on one operon and these can also contain protein-coding genes.
In eukaryotes , a distinction must be made between tRNA genes on the DNA in the cell nucleus and tRNA genes on the DNA of mitochondria (as well as DNA-containing hydrogenosomes ) or plastids such as chloroplasts . In plastids, the genes are organized in operons similar to prokaryotes (see endosymbiont theory ). The equipment with tRNA genes (e.g. 30 tRNA genes in Marchantia polymorpha ) includes all tRNA groups that are required for protein synthesis in plastids.
However, a complete set of tRNA genes is not always available in mitochondria, so that - depending on the species - tRNAs have to be imported from the cytosol into the organelle. While in almost all opisthoconta (e.g. in humans or in yeast ) all mitochondrial tRNA genes are encoded by the mitochondrial DNA, in cnidarians or apicomplexes , for example, most tRNAs have to be imported.
Transcription of the tRNA
Prokaryotes only have one RNA polymerase . This transcribes the reading frame of an operon, so that a polycistronic mRNA is usually produced. If necessary, the tRNAs have to be extracted from this by endonucleolytic cuts in the intercistronic areas.
In eukaryotes, the tRNA is transcribed in the nucleus by RNA polymerase III. In contrast, protein-coding genes are transcribed there by RNA polymerase II. The tRNA in plastids is transcribed by an RNA polymerase, which is similar to that of prokaryotes (see also endosymbiont theory ). This RNA polymerase is also encoded in the plastid genome, while another has to be imported from the cytoplasm . There are two RNA polymerases in the mitochondria, both of which must be imported. They are related to the nucleus-coded plastid enzyme.
In archaea , as in the prokaryotes, there is only one RNA polymerase for transcription. Compared to prokaryotes, however, this is more complex. The size, number of subunits and their amino acid sequences are more comparable to eukaryotic RNA polymerases.
Maturation of the tRNA precursors
The pre-tRNAs resulting from the transcription can contain introns . In bacteria , the splicing of these introns takes place autocatalytically , whereas the introns in eukaryotes and archaea are removed by tRNA splicing endonuclease .
In eukaryotes, both the tRNA synthesis and the first post-transcriptional processing such as the trimming of the 5 'and 3' ends, base modifications and the post-transcriptional addition of the 3 'CCA sequence take place in the nucleus. The export to the cytosol is mediated by proteins such as Los1. In the cytosol, splicing takes place on the outer membrane of mitochondria, if introns are present, and also further modifications until the tRNA has matured.
Origin of the tRNAs
The upper half of the tRNA (from the D-arm and the acceptor strain with the 5'-terminating phosphate unit and the 3'-terminating CCA group) and the lower half (from the T-arm and the anticodon arm) are Units that are independent in structure and function. It is believed that the upper half developed first, with the CCA group originally - i.e. in an early RNA world - marking tRNA-like molecules for the purpose of replication (such a marker is called 'Genomic Tag' in English) ). The lower half could have been created later as an extension, e.g. B. when protein biosynthesis started and the RNA world changed into a ribonucleoprotein world ( RNP world ). Such a scenario is called 'Genomic Tag Hypothesis' in English. In fact, tRNA-like molecules still play an important catalytic role (ergo as ribozymes ) in replication. They can be viewed as molecular (or chemical) fossils that, like the " living fossils ", provide clues about the early course of evolution.
Variations of tRNA forms in the animal kingdom
The “classic” form of the tRNA shown above with a three-armed “clover leaf” structure has been confirmed in previous studies for most of the tRNAs. In some strains, however, heavily modified mitochondrial tRNAs (mt-tRNAs) were found in which various parts of the normal structure were heavily modified or completely lost. It seems unlikely that these are pseudogenes that have become functionless due to mutations . In the case of a tRNA sequence, the D arm is missing in the entire animal kingdom, which suggests that this sequence was already modified in the ancestors of all modern animals. Particularly highly modified mitochondrial tRNA structures have been identified in nematodes and arachnids . Some mt-tRNAs have lost large parts of their usual structures without losing their functionality. Usually, instead of the lost structure, a replacement structure, e.g. B. an additional loop (TV replacement loop) formed. Why the loss of an otherwise highly conserved structure, the mutation of which, according to all assumptions, should almost always be fatal, is possible here as an exception, has not yet been clarified.
See also
literature
- Reginald Garrett, Charles M. Grisham: Biochemistry. (International Student Edition). 4th edition. Cengage Learning Services, 2009, ISBN 978-0-495-11464-2 .
- Peter Karlson, Detlef Doenecke, Jan Koolman, Georg Fuchs, Wolfgang Gerok: Karlsons Biochemie und Pathobiochemie . 15th edition. Georg Thieme, 2005, ISBN 3-13-357815-4 , p. 142.
- Dieter Soll (Ed.), Uttam L. RajBhandary (Ed.), TL Rajbhandary: tRNA: Structure, Biosynthesis, and Function . Asm Press, 1995, ISBN 1-55581-073-X .
- EM Phizicky, AK Hopper: tRNA biology charges to the front. In: Genes Dev . 24 (17), 2010, pp. 1832-1860. PMID 20810645 ; PMC 2932967 (free full text, PDF).
- MA Rubio, AK Hopper: Transfer RNA travels from the cytoplasm to organelles. In: Wiley Interdiscip Rev RNA . 2 (6), 2011, pp. 802-817. PMID 21976284 ; doi: 10.1002 / wrna.93
- Paul Schimmel: The emerging complexity of the tRNA world: mammalian tRNAs beyond protein synthesis . In: Nature Reviews. Molecular Cell Biology . September 6, 2017, doi : 10.1038 / nrm.2017.77 , PMID 28875994 .
- Kunal Chatterjee, Regina T. Nostramo, Yao Wan, Anita K. Hopper: tRNA dynamics between the nucleus, cytoplasm and mitochondrial surface: Location, location, location . In: Biochimica Et Biophysica Acta . November 27, 2017, doi : 10.1016 / j.bbagrm.2017.11.007 , PMID 29191733 .
Individual evidence
- ^ RK Kumar, DR Davis: Synthesis and studies on the effect of 2-thiouridine and 4-thiouridine on sugar conformation and RNA duplex stability. In: Nucleic acids research . Volume 25, Number 6, March 1997, pp. 1272-1280. PMID 9092639 , PMC 146581 (free full text).
- ↑ Yuchen Liu et al .: Biosynthesis of 4-Thiouridine in tRNA in the Methanogenic Archaeon Methanococcus maripaludis. In: The Journal of Biological Chemistry . 287, August 17, 2012, pp. 36683-36692, doi: 10.1074 / jbc.M112.405688
- ^ H. Grosjean, R. Gupta, ES Maxwell: Modified nucleotides in archaeal RNAs. In: P. Blum (Ed.): Archaea: new models for prokaryotic biology. Caister Academic Press, Norfolk, UK 2008, pp. 171-196.
- ↑ SH Kim et al .: Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. In: Science . 185 (149), 1974, pp. 435-440. PMID 4601792 ; doi: 10.1126 / science.185.4149.435 .
- ↑ JD Robertus et al .: Structure of yeast phenylalanine tRNA at 3 Å resolution. In: Nature . 250 (467), 1974, pp. 546-551. PMID 4602655 ; doi: 10.1038 / 250546a0 .
- ↑ a b H. Shi, PB Moore : The crystal structure of yeast phenylalanine tRNA at 1.93 Å resolution: a classic structure revisited. In: RNA . 6 (8), 2000, pp. 1091-1105. PMID 10943889 ; PMC 1369984 (free full text, PDF).
- ^ Reginald Garrett, Charles M. Grisham: Biochemistry. (International Student Edition). 4th edition. Cengage Learning Services, 2009, ISBN 978-0-495-11464-2 , p. 958.
- ↑ K. Horiuchi, M. Harpel, L. Shen, Y. Luo, K. Rogers, R. Copeland: Mechanistic Studies of Reaction Coupling in Glu-tRNAGln Amidotransferase. In: Biochemistry . Volume 40, No. 21, May 2001, pp. 6450-6457, doi: 10.1021 / bi002599l .
- ↑ Katharina Munk (ed.): Pocket textbook Biology: Genetics. 1st edition. Thieme Verlag, 2010, ISBN 978-3-13-144871-2 , p. 188.
- ^ Peter Karlson, Detlef Doenecke, Jan Koolman, Georg Fuchs, Wolfgang Gerok: Karlsons Biochemie und Pathobiochemie. 15th edition. Thieme, 2005, p. 142.
- ^ F. Crick: The origin of the genetic code . In: J Mol Biol . tape 38 , no. 3 , 1968, p. 367-379 , doi : 10.1016 / 0022-2836 (68) 90392-6 , PMID 4887876 .
-
↑ H. Lodish et al .: Molecular Biology of the Cell. 5th edition. WH Freeman, New York 2004, ISBN 0-7167-3136-3 . (Excerpt) (English)
Harvey Lodish et al .: Molecular Cell Biology. Spectrum Academic Publishing House, 2001, ISBN 3-8274-1077-0 . - ↑ a b A. Ambrogelly et al .: Natural expansion of the genetic code. In: Nature Chemical Biology . 3 (1), 2007, pp. 29-35. PMID 17173027 ; doi: 10.1038 / nchembio847 .
- ^ Reginald Garrett, Charles M. Grisham: Biochemistry. (International Student Edition). 4th edition. Cengage Learning Services, 2009, ISBN 978-0-495-11464-2 , p. 959.
- ^ Reginald Garrett, Charles M. Grisham: Biochemistry . (International Student Edition). 4th edition. Cengage Learning Services, 2009, ISBN 978-0-495-11464-2 , p. 977.
- ^ Reginald Garrett, Charles M. Grisham: Biochemistry . (International Student Edition). 4th edition. Cengage Learning Services, 2009, ISBN 978-0-495-11464-2 , p. 966.
- ^ Donald Voet, Judith G. Voet: Biochemistry. Wiley-VCH, 1994, ISBN 3-527-29249-7 , p. 928.
- ^ SE Kolitz, JR Lorsch: Eukaryotic initiator tRNA: finely tuned and ready for action. In: FEBS Letters . 584 (2), January 21, 2010, pp. 396-404 ( doi: 10.1016 / j.febslet.2009.11.047 , PMC 2795131 (free full text). PMID 19925799 ).
- ↑ WormBase : Release Notes - WS187 . January 25, 2008.
- ↑ J. Spieth, D. Lawson: Overview of gene structure. In: WormBook . 2006, pp. 1-10. PMID 18023127 ; PDF (free full text access)
- ↑ LH Hartwell et al .: Genetics: From Genes to Genomes. 2nd Edition. McGraw-Hill, New York 2004, p. 264.
- ^ André Schneider: Mitochondrial tRNA import and its consequences for mitochondrial translation. In: Annu Rev Biochem . 80, 2011, pp. 1033-1053. PMID 21417719 ; doi: 10.1146 / annurev-biochem-060109-092838
- ↑ RJ White: Regulation of RNA polymerases I and III by the retinoblastoma protein: a mechanism for growth control? In: Trends in Biochemical Sciences . 22 (3), 1997, pp. 77-80. PMID 9066256 ; doi: 10.1016 / S0968-0004 (96) 10067-0
- ↑ J. Abelson, CR Trotta, H. Li: tRNA Splicing. In: J Biol Chem 273 (21), 1998, pp. 12685-12688. PMID 9582290 ; PDF (free full text access)
- ^ AK Hopper, DA Pai, DR Engelke: Cellular dynamics of tRNAs and their genes. In: FEBS Lett . 584 (2), 2010, pp. 310-317. PMID 19931532 ; PMC 2818515 (free full text, PDF).
- ^ Nancy Maizels, Alan M. Weiner: The Genomic Tag Hypothesis - What Molecular Fossils Tell Us about the Evolution of tRNA. In: The RNA World. 2nd Edition. Cold Spring Harbor Laboratory Press, 1999, ISBN 0-87969-561-7 . (PDF)
- ↑ DR Wolstenholme: Animal mitochondrial DNA: structure and evolution. In: DR Wolstenholme, KW Jeon (Ed.): Mitochondrial genomes. Academic Press, New York 1992, pp. 173-216.
- ^ Y. Watanabe et al .: Primary and higher order structures of nematode (Ascaris suum) mitochondrial tRNAs lacking either the T or D stem. In: Journal of Biological Chemistry . 269 (36), pp. 22902-22906. PMID 8077242 ; PDF (free full text access)
- ↑ Susan E. Masta, Jeffrey L. Boore: Parallel Evolution of Truncated Transfer RNA Genes in Arachnid Mitochondrial Genomes. In: Molecular Biology and Evolution . 25 (5), 2008, pp. 949-959. PMID 18296699 ; PDF (free full text access)
Web links
- The tRNA modification database ( Memento from April 7, 2013 in the Internet Archive ) - Comprehensive database on modified bases in tRNAs
- Mammalian mitochondrial tRNA database
- The Genomic tRNA Database (GtRDB)








