Secondary structure
In biochemistry, regular local structural elements of macromolecules are called secondary structures . The focus is on the polymer backbone, also known as the backbone . The conformation of the side chains and their relationship to other elements are disregarded.
The representation of the secondary structure of a polymer offers a better overview than the representation of its complete molecular structure and at the same time gives a much more precise insight into the actual structure than the representation in the Pauling notation or the Fischer projection .
The secondary structure emerges from the primary structure . Hydrogen bonds between the atoms are responsible for the topological arrangement of the atoms in space . The form of the polymerisation, for example in the case of polysaccharides , can also influence the secondary structure.
The tertiary structure and the quaternary structure represent superordinate structural levels. The division into a hierarchy into primary structure, secondary structure, tertiary structure and quaternary structure was proposed in 1952 by Kaj Ulrik Linderstrøm-Lang .
Secondary structure of proteins

In proteins, the secondary structure is determined by hydrogen bonds between the CO and NH groups of the peptide backbone. This results in a vast number of conformational possibilities for each protein. However, it turns out that some motifs can be found again and again. These are called secondary structural elements .
A distinction is made between:
- α-helix
- π helix
- 3 10 -Helix
- left-handed α-chain of the collagens
- β-sheet
- β loop
- β-helix
- Random coil : Areas that do not have a defined secondary structure
With the exception of the beta loops and random coils , these areas are characterized by the fact that the only two possible angles of rotation ψ and φ of the peptide backbone are fixed in them and repeat themselves periodically over the length of the secondary structural element. In the Ramachandran plot , the possible secondary structures are shown as a function of the associated ψ / φ angle pairs, in the Janin plot the dihedral angles of the amino acid side chains (χ 1 and χ 2 ). The secondary structural elements are energetically stabilized by the hydrogen bridges within the peptide backbone . Depending on the type of secondary structural element, certain amino acid side chains can have a destabilizing effect on its structure.

In addition, there is another interaction through the π * orbital of the C = O carbon with one of the two lonepairs of the subsequent C = O oxygen. This leads to a pyramidalization of the otherwise planar amides .
The secondary structure of a protein consists of various secondary structural elements. The next higher level of classification are structural motifs and also protein domains . The complete protein structure (i.e. the sequence or arrangement of the secondary structural elements) is called the tertiary structure. It is characteristic of every protein and is essential for biological function. During or after the translation of an RNA molecule, the secondary structure and the higher structural levels of the protein are formed. This process, called protein folding , is common in most proteins. a. supported by chaperones , but sometimes also happens spontaneously with small proteins.
Secondary structure of nucleic acids
Also, nucleic acids , ie DNA and RNA can form secondary structures. The prerequisite for this is that the nucleic acid molecule is initially available as a single strand. Complementary sections of the strand can then form hydrogen bonds, which leads to the formation of intramolecular base pairs with a double-stranded structure. Non-complementary sections remain single-stranded.
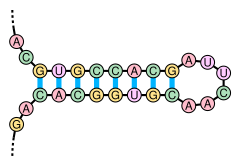
The paired portions of the double-stranded region present, stem ( Stem ) called, can have a longer single-stranded portion therebetween as a loop ( loop include) and as a secondary stem loop structure ( stem-loop ) form. Secondary structures of nucleic acids with a small loop, so a short single-stranded intermediate portion is referred to on the basis of their shape as a hairpin structure ( hairpin ). The opposing pairing of complementary base sequences is made possible in a special way by palindromic sequences in the nucleotide sequence of a nucleic acid strand.
The secondary structures have important functions in the regulation of transcription . They can serve as primers (see telomerase ) or be a prerequisite for the enzymatic activity of the ribosomes (see rRNA ).
RNA structure prediction algorithms
Individual evidence
- ↑ Entry on secondary structure . In: IUPAC Compendium of Chemical Terminology (the “Gold Book”) . doi : 10.1351 / goldbook.S05530 .
- ^ Hermann J. Roth, Christa E. Müller, Gerd Folkers: Stereochemie und Arzneimittel , Wissenschaftliche Verlagsgesellschaft Stuttgart, 1998, pp. 240–244, ISBN 3-8047-1485-4 .
- ^ Horton, Robert et al .: Biochemie , 4th edition, Pearson Studium, Munich (2008), p. 152, ISBN 978-3-8273-7312-0 .
