Ribosome
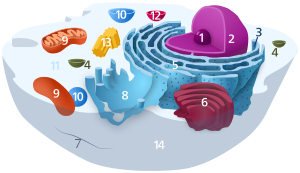
1. Nucleolus (nuclear body)
2. Cell nucleus (nucleus)
3. Ribosomes
4. Vesicle
5. Rough (granular) ER (ergastoplasm)
6. Golgi apparatus
7. Cytoskeleton
8. Smooth (agranular) ER
9 . mitochondria
10. lysosome
11. cytoplasm (with cytosol and cytoskeleton )
12. peroxisomes
13. centrioles
14 cell membrane
Ribosomes are the macromolecular complexes in cells that make proteins. Here, the nucleotide sequence (base sequence) of a messenger ribonucleic acid single strand (mRNA) is translated into the amino acid sequence of the polypeptide chain of a protein . This conversion of the information stored in the RNA into a sequence of linked amino acids is called translation ( Latin for translation ) and is a central component of protein biosynthesis in all living things . The translation rule in effect here is called the genetic code . The translation takes place in the cell after the genetic information of a gene , which is stored in the sequence of base pairs of the DNA double strand, has been rewritten into the sequence of the mRNA single strand .
Ribosomes are made up of ribosomal RNA , English ribonucleic acid (rRNA) and proteins (rProtein, also r-protein) and are found in the cytoplasm as well as in cell organelles which, due to their endosymbiotic origin, have their own machinery for protein biosynthesis, such as the mitochondria and chloroplasts .
Structure and types
Ribosomes are granular particles with a diameter of about 20-25 nm. They consist of about two thirds of RNA ( rRNA ) and one third of ribosomal proteins. In all organisms they are made up of two differently sized and functionally different subunits. The mass of the ribosomes is characterized by their sedimentation behavior, which is given in Svedberg units (S). During translation, they assemble into a functional complex, with the large subunit linking the amino acids into a chain in protein biosynthesis (peptidyl transferase activity), and the small subunit being responsible for mRNA recognition. Both subunits consist of proteins and rRNA, whereby the proteins are responsible for cohesion and correct positioning, while the actual reactions are carried out by the rRNAs. In eukaryotes, both subunits are formed in the nucleoli within the cell nuclei and are then passed through the nuclear pores into the cytoplasm.
Prokaryotic ribosomes
| Ribosome | Subunit | rRNAs | r proteins |
|---|---|---|---|
| 70 p | 50S | 23S (2904 nt ) | 31 |
| 5S (120 nt) | |||
| 30S | 16S (1542 nt) | 21st |
The number of ribosomes per cell in prokaryotes is in the order of 10,000, for example a single E. coli bacterium has around 20,000 ribosomes. The ribosomes have a sedimentation coefficient of 70S and a molar mass of about 2.5 MDa . At magnesium concentrations below 1 mmol / l, the 70S ribosome breaks down into a 50S and a smaller 30S subunit. The 30S subunit (0.9 MDa) is composed of 21 different ribosomal proteins and one 16S ribosomal RNA (16S rRNA). The 50S subunit (1.6 MDa) contains 31 different proteins and two rRNAs (23S and 5S rRNA).
The proteins of the small subunit are marked with "S" ( English small , small '), those of the large subunit with "L" (English large , large'). Their amino acid sequences have nothing in common, but they are rich in positively charged amino acids such as L - lysine or L - arginine . This allows a better interaction with the negatively charged rRNAs. The largest bacterial ribosomal protein is S1 with 61.2 kDa and 557 amino acids, the smallest is L34 with 5.4 kDa and 34 amino acids.
Eukaryotic ribosomes
In eukaryotic cells, the ribosomes are located in the cytoplasm (not in the karyoplasm of the cell nucleus). In addition, special ribosomes also occur in some organelles if they have their own DNA:
- in the mitochondria (or alternatively in hydrogenosomes if with DNA), as well
- in (almost all) chloroplasts and other plastids , if - as in plants - present.
Cytosolic ribosomes
| Ribosome | Subunit | rRNAs | r proteins |
|---|---|---|---|
| 80S | 60S | 28S (4718 nt) | 49 |
| 5.8S (160 nt) | |||
| 5S (120 nt) | |||
| 40S | 18S (1874 nt) | 33 |
The number of cytosolic ribosomes per cell is estimated to be between 10 5 and over 10 7 , which means that eukaryotic cells have more ribosomes than prokaryotic cells. The number depends on the cell type, namely on the protein synthesis rate of the cell. The number of ribosomes in liver cells is particularly high. In addition, the eukaryotic ribosomes of the cytosol are also larger, they have a diameter of about 25 nm. They have a molar mass of about 4.2 MDa, the sedimentation coefficient is 80S. For the large subunit it is 60S (2.8 MDa) and for its small subunit it is 40S (1.4 MDa). In mammals, the small subunit consists of 33 proteins and one rRNA (18S rRNA), the large subunit consists of 49 proteins and three rRNAs (28S, 5.8S and 5S). Cytosolic ribosomes of higher eukaryotes are more complex than the lower eukaryotes. The 28S rRNA in baker's yeast is 3,392 nucleotides long, whereas in mammals such as rats it is 4,718 nucleotides. The 18S rRNA is also smaller in baker's yeast than in the rat (1799 compared to 1,874 nucleotides).
The actual catalytic function is carried out by the rRNA, whereas the proteins are located at the edge of the ribosome. In addition to the free cytoplasmic ribosomes, eukaryotes also have membrane-bound ribosomes that are bound to the membrane of the rough endoplasmic reticulum (ER) (see below). The formation of the ribosomal subunits takes place in the nucleolus . Cells with a high rate of protein synthesis therefore have particularly well-developed nucleoli. Free and membrane-bound ribosomes have the same structure and can switch between functions.
The notation of eukaryotic ribosomal proteins is not entirely uniform. In baker's yeast, proteins of the large subunit are called “Rpl”, those of the small ones are called “Rps”. The capitalization RPL or RPS is also used for the corresponding mammalian proteins.
Mitoribosomes and Plastoribosomes
The ribosomes from mitochondria and chloroplasts are similar to the prokaryotic ribosomes, which supports the endosymbiont hypothesis . The mitochondrial ribosomes of humans and other mammals are composed of many proteins, 21 of which are only found in mitochondria, and produce only mitochondrial membrane proteins .
80S ribosomes in complex plastids
In contrast to this, the complex plastids of chlorine arachniophytes, for example, with an additional nucleus ( nucleomorph ) can contain their own eukaryotic 80S ribosomes. The complex plastids are interpreted as the result of a secondary endosymbiosis ( secondary plastids ).
Free and membrane-bound ribosomes
Ribosomes can be differentiated in eukaryotic cells according to their place of synthesis. Free ribosomes are scattered in the cytoplasm and produce proteins that mostly also perform their task in the cell plasma. Membrane-bound ribosomes are attached to the membrane of the endoplasmic reticulum. The proteins synthesized there are guided into the lumen of the endoplasmic reticulum by means of cotranslational protein transport . Membrane-bound ribosomes are often found in secretion-forming cells such as z. B. in the pancreas.
functionality

The functioning of the ribosome during translation can be characterized by the three-digit model. According to this, the ribosome has three tRNA binding sites, the A (aminoacyl), P (peptidyl) and E (exit) sites. During the elongation cycle, the ribosome oscillates between two states, the pre- and post-translational state, whereby two of the three tRNA binding sites are occupied by a tRNA. In the pretranslational state, the A and P sites are occupied, the P site carrying the tRNA with the polypeptide chain and the A site being occupied by the newly added aminoacyl-tRNA. In the ribosome, the polypeptide chain is now transferred from the P-site tRNA to the A-site tRNA using peptidyl transferase. The ribosome then switches to the post-translational state and moves three bases on the mRNA, whereby the previous A-site tRNA becomes the P-site tRNA and the now empty former P-site tRNA via the E-site (Exit ) is channeled out of the ribosome. A translocase (EF-G) is involved.
The two main states of the ribosome (pre- and post-translational) are separated from each other by a high activation energy barrier. The central role of the two elongation factors is to lower this energy barrier and thus to put the ribosome in the other state.
Sometimes several prokaryotic ribosomes form a string of pearls on the same mRNA molecule to form a polysome .
After a peptide has been linked in the ribosome, it travels through a ribosomal tunnel. This consists largely of rRNA and emerges from the large ribosomal subunit. It is approximately 100 Å (10 nm) long and has an average diameter of 15 Å (1.5 nm). At its narrowest point the channel is bounded by two conserved ribosomal proteins, L4e and L22.
Ribophagy
The breakdown of ribosomes is not yet fully understood. It is usually initiated when there is a lack of nutrients. For bacteria such as E. coli , it has been suggested that intact 70S ribosomes first break down into both subunits. Under deficiency conditions, the translation in the cell is shut down so that many ribosomes are inactive. The two subunits are much more sensitive to ribonucleases (RNases) than an intact ribosome, as they offer a larger target. Thereafter, exonucleases could also degrade the ribosomal RNA further.
For baker's yeast, a eukaryote, an autophagy route called "ribophagy" has been proposed. This is based on the terms mitophagy (breakdown of mitochondria), pexophagy (breakdown of peroxisomes) and reticulophagy (breakdown of the endoplasmic reticulum). When there is a lack of nutrients, yeast break down ribosomes in a way that begins similarly to that of prokaryotes. First, the two sub-units are separated. A ubiquitin ligase then removes ubiquitin on the 60S subunit, which is then transported to the vacuole in a vesicle . This seems paradoxical at first, since ubiquitin is a general degradation signal for most proteins. The authors suggested that a ubiquitin ligase initially marks the 60S subunit for the degradation pathway, but that the process can only finally take place through the ubiquitin protease.
Structure elucidation
Ribosomes were discovered by the researcher Albert Claude in the middle of the 20th century. In 1940 he had identified RNA-containing granules from the cytosol of animal cells that were smaller than mitochondria with the help of dark field microscopy . He called these “microsomes”, later analyzes showed that they were complexes of phospholipids and ribonucleic proteins. Today, fragments of the ER are called microsomes . In 1955, through advances in electron microscopy , George Emil Palade succeeded in clearly identifying those “microsomes” as components of a cell and not just as artifacts from cell debris. There was increasing evidence that these ribonucleic protein particles had something to do with translation. In 1959, E. coli also provided evidence that ribosomes are necessary for the biosynthesis of polypeptides.
In 1958 Richard B. Roberts took up the suggestion in a symposium to change the name "microsome" or "microsome particle" to the better-sounding and simple name - according to Roberts - "ribosome". This abbreviation refers to the type of particles, complexes of RNA and proteins (ribonucleoparticles). The term "ribosome" was able to gain acceptance and is used in everyday language.
Because of their size, it has only recently been possible to obtain high-resolution structures from ribosomes, although the coarse molecular structure has been known since the 1970s. Some details of ribosomal proteins could be elucidated by means of affinity labeling and chemical crosslinking . At the end of 2000 the 50S subunit of the archaeon Haloarcula marismortui was elucidated for the first time with a resolution of 2.4 Å . Individual molecules can be dissolved in this resolution. At the same time, the structures of the small ribosomal subunit from Thermus thermophilus were published with an atomic resolution of 3 Å. Since no structural data for the complete ribosome were available at this point in time, the available data were used to reconstruct the prokaryotic ribosome.
While the A and P positions had been known for a long time, the E position was not discovered until 1981 ( Knud Nierhaus and colleagues, Alpha-epsilon theory of the binding of t-RNA in the ribosome).
In 2005, the crystallographic structural data of an intact ribosome from E. coli were presented for the first time with a resolution of 3.5 Å. Almost at the same time, another research group was able to present a structure that was obtained with the help of cryoelectron microscopy. The resolution was comparatively low at over 10 Å, but showed a snapshot of the translation at the translocon.
Later, more and more structural data of (prokaryotic) ribosomes that had just bound mRNAs or tRNAs were published and thus provided a better insight into the processes of translation.
No comparable structural data are yet available for the eukaryotic ribosome (80S). A three-dimensional reconstruction is possible from the data collected from cryoelectron microscopy , X-ray crystallography of individual ribosomal components and homology comparisons with prokaryotic ribosomes.
Thomas A. Steitz , Ada Yonath and Venkatraman Ramakrishnan received the Nobel Prize in Chemistry in 2009 for their work on structure elucidation .
origin
The origin of the ribosomes is believed to be in the RNA world , in which a self-replicating complex only developed the ability to synthesize proteins later, when sufficient amino acids were available. The catalytic abilities of RNA ( ribozyme ) are a central component of the RNA world hypothesis. Research suggests that these ribosome precursors, built entirely from rRNA , may have developed the ability to form peptide bonds . In addition, there is strong evidence that original ribosomes were self-replicating complexes in which the rRNA served informational, structural, and catalytic purposes, as it may have encoded tRNA and proteins for ribosomal self-replication. The hypothetical DNA-free cellular organisms that were equipped with such self-replicating RNA are called ribocytes .
As amino acids gradually accumulated in the RNA world under conditions that were still prebiotic , their interaction with the catalytic RNA could have increased both its scope and its efficiency. The selective pressure to build proteins into the self-replicating mechanisms of the ribosomes could have been the driving force for the evolution of the ribosomes from an originally self-replicating machine to its current form as a translation machine, as this would have increased the capacity of self-replication.
Origin of DNA
The storage of the genome in the form of the DNA double helix appears to be a later ingredient. DNA replication and transcription are so different in bacteria on the one hand, and archaea and eukaryotes ( Neomura ) on the other hand, that the assumption of a common origin ( homology ) seems improbable. Instead, these two groups - based on primitive cellular organisms with ribosomes - could each have acquired the ability to store genetic information in DNA, presumably with the help of DNA viruses . According to this assumption, the DNA viruses had previously developed from the more original RNA viruses in order to better protect their genome from attacks by the host cells, which meant the end of the pure RNA world.
literature
- Donald Voet and Judith G. Voet: Biochemistry . Wiley-VCH 1994; ISBN 3-527-29249-7 ; P. 917ff.
- Alexander S. Spirin: Ribosomes (Cellular Organelles) . Springer, Berlin 1999; ISBN 0-306-46145-5
- Reginald Garrett and Charles M. Grisham: Biochemistry . (International Student Edition). Cengage learning services; 4th edition 2009; ISBN 978-0-495-11464-2
- S. Klinge et al . (2012): Atomic structures of the eukaryotic ribosome . In: Trends Biochem Sci . 37 (5); 189-198; PMID 22436288 ; doi: 10.1016 / j.tibs.2012.02.007
- DN Wilson and JH Doudna Cate (2012): The structure and function of the eukaryotic ribosome . In: Cold Spring Harb Perspect Biol . 4 (5); 1-17; PMID 22550233 ; PDF (free full text access)
- S. Melnikov et al . (2012): One core, two shells: bacterial and eukaryotic ribosomes . In: Nat Struct Mol Biol . 19 (6); 560-567; PMID 22664983 ; doi: 10.1038 / nsmb.2313
Web links
- Proteopedia: Ribosomes
- Ribosomal Database (database on ribosomal proteins)
- Video: explanation of ribosome structure and its discovery
- Video: Protein Biosynthesis at the Ribosome (Kenyon College, English) ( MP4 ; 39.8 MB)
Individual evidence
- ^ Hans G. Kloepfer: Structure and function of ribosomes . In: Chemistry in Our Time . tape 7 , no. 2 , 1973, p. 49-58 , doi : 10.1002 / ciuz.19730070204 .
- ↑ Salini Konikkat: Dynamic Remodeling Events Drive the Removal of the ITS2 spacer sequence During Assembly of 60S ribosomal subunits in S. cerevisiae. Carnegie Mellon University Dissertations, Feb. 2016.
- ↑ Elmar W. Weiler, Lutz Nover: General and molecular botany . Georg Thieme Verlag, Stuttgart 2008, ISBN 978-3-13-152791-2 , p. 532 ( limited preview in Google Book search).
- ↑ Jesus de la Cruz, Katrin Karbstein, John L. Woolford, Jr .: Functions of Ribosomal Proteins in Assembly of Eukaryotic Ribosomes In Vivo . In: Annual review of biochemistry . tape 84 , 2015, p. 93-129 , doi : 10.1146 / annurev-biochem-060614-033917 , PMID 25706898 , PMC 4772166 (free full text).
- ^ A b Reginald Garrett and Charles M. Grisham: Biochemistry . (International Student Edition). Cengage learning services; 4th edition 2009; ISBN 978-0-495-11464-2 ; P. 962.
- ^ Reginald Garrett and Charles M. Grisham: Biochemistry . (International Student Edition). Cengage learning services; 4th edition 2009; ISBN 978-0-495-11464-2 ; P. 965.
- ↑ Helmut Plattner and Joachim Hentschel: Cell Biology . Thieme, Stuttgart; 3., rework. Edition 2006; ISBN 3-13-106513-3 ; P. 181.
- ^ Reginald Garrett and Charles M. Grisham: Biochemistry . (International Student Edition). Cengage learning services; 4th edition 2009; ISBN 978-0-495-11464-2 ; P. 964.
- ^ A. Brown, A. Amunts, XC Bai, Y. Sugimoto, PC Edwards, G. Murshudov, SH Scheres, V. Ramakrishnan: Structure of the large ribosomal subunit from human mitochondria. In: Science. Volume 346, number 6210, November 2014, pp. 718–722, doi: 10.1126 / science.1258026 . PMID 25278503 .
- ^ BJ Greber, D. Boehringer, M. Leibundgut, P. Bieri, A. Leitner, N. Schmitz, R. Aebersold, N. Ban: The complete structure of the large subunit of the mammalian mitochondrial ribosome. In: Nature . Volume 515, number 7526, November 2014, pp. 283–286, doi: 10.1038 / nature13895 . PMID 25271403 .
- ↑ Shigekatsu Suzuki, Shu Shirato, Yoshihisa Hirakawa, Ken-Ichiro Ishida: Nucleomorph Genome Sequences of Two Chlorarachniophytes, Amorphochlora amoebiformis and Lotharella vacuolata . In: Genome Biology and Evolution . tape 7 , no. 6 , 2015, ISSN 1759-6653 , p. 1533–1545 , doi : 10.1093 / gbe / evv096 , PMID 26002880 , PMC 4494063 (free full text).
- ^ Zundel, MA. et al . (2009): Initiation of ribosome degradation during starvation in Escherichia coli . In: RNA 15 (5); 977-783; PMID 19324965 ; PDF (free full text access, English).
- ^ Kraft, C. et al . (2008): Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p / Bre5p ubiquitin protease . In: Nat Cell Biol . 10 (5); 602-610; PMID 18391941 ; doi: 10.1038 / ncb1723
- ↑ Kim, I. et al . (2007): Selective degradation of mitochondria by mitophagy . In: Arch Biochem Biophys . 462 (2); 245-253; PMID 17475204 ; PMC 2756107 (free full text)
- ↑ Dunn, WA. Jr. et al . (2005): Pexophagy: the selective autophagy of peroxisomes . In: Autophagy 1 (2); 75-83; PMID 16874024 ; PDF (free full text access, English).
- ↑ Klionsky, DJ. et al . (2007): How shall I eat thee ? In: Autophagy 3 (5); 413-416; PMID 17568180 ; PDF (free full text access, English).
- ↑ Donald Voet and Judith G. Voet: Biochemistry . Wiley-VCH 1994; ISBN 3-527-29249-7 ; P. 917
- ↑ Alexander S. Spirin: Ribosomes (Cellular Organelles) . Springer, Berlin 1999; ISBN 0-306-46145-5 , p. 47.
- ↑ Claude, A. (1940): Particulate components of normal and tumor cells . In: Science 91 (2351); 77-78: PMID 17783332 ; doi: 10.1126 / science.91.2351.77
- ↑ Palade, GE. (1955): A small particulate component of the cytoplasm . In: J Biophys Biochem Cytol . 1 (1); 59-68; PMID 14381428 ; PMC 2223592 (free full text)
- ↑ McQuillen, K., Roberts, RB. and Britten, RJ. (1959): SYNTHESIS OF NASCENT PROTEIN BY RIBOSOMES IN ESCHERICHIA COLI. In: Proc Natl Acad Sci USA 45 (9): 1437-1447; PMID 16590524 ; PMC 222733 (free full text)
- ↑ Richard B. Roberts: Microsomal Particles and Protein Synthesis . London, Pergamon Press 1958; Full text access (English).
- ↑ Czernilofsky, AP., Kurland, CG. and Stöffler, G. (1975): 30S ribosomal proteins associated with the 3'-terminus of 16S RNA. In: FEBS Lett . 58 (1); 281-284; PMID 1225593 .
- ↑ Ban, N. et al . (2000). The complete atomic structure of the large ribosomal subunit at 2.4 Angstrom resolution . In: Science 289 (5481); 905-920; PMID 10937989 ; doi: 10.1126 / science.289.5481.905
- ↑ Schluenzen, F. et al . (2000): Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution . In: Cell 102 (5); 615-623; PMID 11007480
- ↑ Wimberly, BT., Et al . (2000): Structure of the 30S ribosomal subunit . In: Nature . 407 (6802); 327-339; PMID 11014182 ; doi: 10.1038 / 35030006
- ↑ Yusupov, MM. Et al . (2001): Crystal structure of the ribosome at 5.5 Angstrom resolution . In: Science. 292 (5518); 883-896. PMID 11283358 ; doi: 10.1126 / science.1060089
- ↑ Schuwirth, BS et al . (2005): Structures of the bacterial ribosome at 3.5 A resolution . In: Science 310 (5749): 827-834; PMID 16272117 ; doi: 10.1126 / science.1117230 .
- ↑ Mitra, K. et al . (2005): Structure of the E. coli protein-conducting channel bound to a translating ribosome . In: Nature. 438 (7066); 318-324; PMID 16292303 ; PMC 1351281 (free full text).
- ↑ Selmer, M. et al (2006): Structure of the 70S ribosome complexed with mRNA and tRNA . In: Science 313 (5795): 1935-1942; PMID 16959973 .
- ↑ Korostelev, A., Trakhanov, S., Laurberg, M. and Noller, HF (2006): Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements . In: Cell 126 (6); 1065-1077; PMID 16962654 .
- ↑ Gilbert, RJ et al . (2004): Three-dimensional structures of translating ribosomes by Cryo-EM . In: Mol Cell 14 (1): 57-66; PMID 15068803 .
- ↑ Stark, H. (2002): Three-dimensional electron cryomicroscopy of ribosomes . In: Curr Protein Pept Sci 3 (1): 79-91; PMID 12370013 .
- ↑ Spahn, CM, et al . (2001): Structure of the 80S ribosome from Saccharomyces cerevisiae - tRNA-ribosome and subunit-subunit interactions . In: Cell 107 (3): 373-386; PMID 11701127 .
- ↑ Heena Khatter, Alexander G. Myasnikov, S. Kundhavai Natchiar, Bruno P. Klaholz: Structure of the human 80S ribosome. In: Nature. 520, 2015, p. 640, doi: 10.1038 / nature14427 .
- ^ Nobel Prize in Chemistry 2009 .
- ↑ a b H. F. Noller: Evolution of protein synthesis from an RNA world . In: Cold Spring Harbor Perspectives in Biology . 4, 2012, pp. 1 – U20. doi : 10.1101 / cshperspect.a003681 . PMC 3312679 (free full text).
- ↑ ER Dabbs: Structure, Function, and Genetics of the ribosome . Ed .: B. Hardesty, G. Kramer. Springer, New York 1986, Chapter 43: Mutant Studies on the Prokaryotic Ribosome , pp. 733-748 , doi : 10.1007 / 978-1-4612-4884-2_43 .
- ↑ HF Noller, V. Hoffarth, L. Zimniak: Unusual resistance of peptidyl transferase to protein extraction procedures . In: Science . tape 256 , no. 5062 , June 1992, p. 1416-1419 , doi : 10.1126 / science.1604315 , PMID 1604315 .
- ↑ M. Nomura, S. Mizushima, M. Ozaki, P. Trau, CV Lowry: Structure and function of ribosomes and their molecular components . In: Cold Spring Harbor Symposium of Quantitative Biology . 34, 1969, pp. 49-61. doi : 10.1101 / sqb.1969.034.01.009 .
- ^ A b M. Root-Bernstein, R. Root-Bernstein: The ribosome as a missing link in the evolution of life . In: Journal of Theoretical Biology . 367, 2015, pp. 130–158. doi : 10.1016 / j.jtbi.2014.11.025 . PMID 25500179 .
- ↑ M. Yarus: Primordial genetics: phenotype of the ribocyte . In: Annu. Rev. Genet. . 36, 2002, pp. 125-51. doi : 10.1146 / annurev.genet.36.031902.105056 . PMID 12429689 .
- ↑ G. Caetano-Anolles, MJ Seufferheld: The coevolutionary roots of biochemistry and cellular organization challenge the RNA world paradigm . In: Journal of Molecular Microbiology and Biotechnology . 23, 2013, pp. 152-177. doi : 10.1159 / 000346551 .
- ^ R. Saladino, G. Botta, S. Pino, G. Costanzo, E. Di Mauro: Genetics first or metabolism first? The formamide clue . In: Chemical Society Reviews . 41, 2012, pp. 5526-5565. doi : 10.1039 / c2cs35066a .
- ↑ P. Forterre : Evolution - The true nature of viruses. In: spectrum. August 2017, p. 37 (online article from July 19, 2017).
- ↑ P. Forterre: Three RNA cells for ribosomal lineages and three DNA viruses to replicate their genomes: a hypothesis for the origin of cellular domain . In: Proc. Natl. Acad. Sci. USA . 103, March 2006, pp. 3669-74. doi : 10.1073 / pnas.0510333103 . PMID 16505372 . PMC 1450140 (free full text).
- ↑ C. Zimmer: Did DNA come from viruses? . In: Science . 312, No. 5775, May 2006, pp. 870-2. doi : 10.1126 / science.312.5775.870 . PMID 16690855 .
