Nucleotides
The building blocks of nucleic acids both in strands of ribonucleic acid (RNA or German RNS) and deoxyribonucleic acid (DNA or German DNA) are referred to as nucleotides , also known as nucleotides (abbreviated nt ) . A nucleotide is made up of a base , a sugar and a phosphate component.
While nucleosides only consist of base and sugar, nucleotides also contain phosphate groups. Differences between individual nucleotide molecules can therefore exist in the nucleobase , the monosaccharide and the phosphate residue.
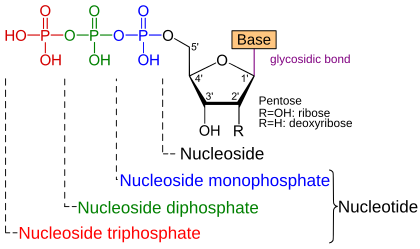
A nucleoside is the combination of a nucleic base (base) with a simple sugar , a pentose . Its 2 'radical (R) is a hydroxyl group (OH-) in the case of ribose , but hydrogen (H-) in the case of deoxyribose . In the case of a nucleotide , the 5'-OH group of the pentose of a nucleoside is esterified with a phosphate residue. A nucleoside triphosphate (NTP) has three phosphate groups that form acid anhydride bonds with one another. Adenosine triphosphate (ATP) is present with adenine as base and ribose as saccharide. In the case of artificial dideoxynucleotides , the 3'-OH group has also been replaced by an H atom.
Nucleotides are not only linked as monophosphates (NMP) in the information-carrying macromolecules of nucleic acids. They also have various other functions for the regulation of life processes in cells . Triphosphates (NTP) such as adenosine triphosphate (ATP) play a central role in the transfer of energy between metabolic pathways, as a cofactor for the activity of enzymes , for transport through motor proteins or the contraction of muscle cells. Guanosine triphosphate (GTP) binding ( G- ) proteins transmit signals from membrane receptors ; cyclic adenosine monophosphate (cAMP) is an important intracellular messenger substance .
Structure of a nucleotide
A nucleotide is made up of three components:
- Base - one of the five nucleobases , namely adenine (A), guanine (G), cytosine (C), thymine (T) or uracil (U)
- Sugar - a monosaccharide ( simple sugar ) with 5 C - atoms , as the furanose ring present pentose , namely ribose ( D -Ribofuranose) or deoxyribose (2-deoxy- D -ribofuranose)
- Phosphate - a residue with at least one phosphate group

The base is usually linked to the sugar via an N- glycosidic bond , the sugar to the phosphate via an ester bond ; If more than one phosphate group is attached, these are linked to one another via phosphoric acid anhydride bonds . In addition, nucleotides naturally occur in which sugar and base are linked via a C-glycosidic bond, for example pseudouridine (Ψ) in transfer RNA .
In the DNA the four nucleobases A, G, C, T are used, in the RNA the four bases A, G, C, U; Instead of the thymine in DNA, uracil is used in RNA. The individual nucleotides differ in the base and the sugar, the eponymous pentose, which is deoxyribose in DNA and ribose in RNA .
Linking of nucleotides to nucleic acids
The macromolecule of a DNA or an RNA is composed of four different types of nucleotides, which are linked by covalent bonds to the strand of the polymeric biomolecule, a polynucleotide . The reaction taking place here is a condensation reaction . A pyrophosphate residue is split off from the monomeric nucleoside triphosphates , so that the monosaccharides of the nucleotides are each coupled to one another via a phosphate group that connects the C5 'atom of the next with the C3' atom of the preceding pentose.

The single strand of a nucleic acid is built up with this pentose phosphate backbone , i.e. with deoxyribose phosphate in a DNA. In the case of a DNA double strand, the bases of the nucleotides of one DNA single strand are opposite the bases of the nucleotides of the other single strand; their phosphate deoxyribose backbone thus points outwards.
Typically, a (smaller) pyrimidine base (T, C) and a (larger) purine base (A, G) each form a pair. The base pairs from T and A and from C and G are referred to as complementary : Opposite a nucleotide that contains cytosine as the base, there is usually a nucleotide with guanine as the base (and vice versa); the same applies to the base pair of adenine and thymine. The opposite bases of the nucleotides of two strands are connected to one another in the DNA double helix via hydrogen bonds . There are three between bases G and C and only two between A and T.
This base-pairing mechanism not only allows the formation of DNA helices. By assigning the complementary base to each of the bases of a single strand, it is also possible to rebuild a complementary single strand. This happens, for example, during replication with the help of a DNA polymerase .
RNA molecules are also made up of nucleotides, with the difference that here ribose is used as a monosaccharide instead of deoxyribose, and that uracil is used as a base instead of thymine. The small differences in the structure of single-stranded RNA and DNA molecules do not prevent them from forming hydrogen bonds between complementary bases as well. Complementary base pairings are also possible within the same strand molecule. For example, certain sections of an RNA molecule can lay on top of one another and fold into hairpin structures . Multiple loop formations are also possible, even typical for the formation of the trefoil structure of tRNA molecules. Although RNA can also be double-stranded, also as a helix, most of the biologically active RNA molecules consist of a single strand.
Three interconnected nucleotides represent the smallest unit of information that is available in DNA or in RNA for coding the genetic information. This information unit is called a codon .
Nucleotides as mono-, di- and triphosphates
Nucleotides consist of a nucleoside and a residue of one, two or three phosphate groups.
- Nucleobase + pentose = nucleoside
- Nucleobase + pentose + 1 phosphate group = nucleotide ( nucleoside monophosphate , NMP)
- Nucleobase + pentose + 2 phosphate groups = nucleotide ( nucleoside diphosphate , NDP)
- Nucleobase + pentose + 3 phosphate groups = nucleotide ( nucleoside triphosphate , NTP)
Ribose as sugar
| Nucleobase | Nucleoside | Nucleotide | Structural formula | ||
|---|---|---|---|---|---|
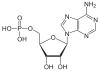
| Adenine | A. | Adenosine | Adenosine monophosphate | AMP |
|
| Adenosine diphosphate | ADP |

|
|||
| Adenosine triphosphate | ATP |

|
|||
| Guanine | G | Guanosine | Guanosine monophosphate | GMP |

|
| Guanosine diphosphate | GDP |

|
|||
| Guanosine triphosphate | GTP |
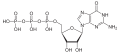
|
|||
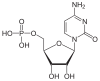
| Cytosine | C. | Cytidine | Cytidine monophosphate | CMP |
|
| Cytidine diphosphate | CDP |

|
|||
| Cytidine triphosphate | CTP |
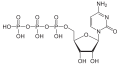
|
|||
| Uracil | U | Uridine | Uridine monophosphate | UMP |

|
| Uridine diphosphate | UDP |
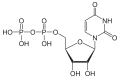
|
|||
| Uridine triphosphate | UTP |
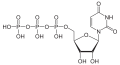
|
|||
Deoxyribose as a sugar
| Nucleobase | Nucleoside | Nucleotide | Structural formula | ||
|---|---|---|---|---|---|
| Adenine | A. | Deoxyadenosine | Deoxyadenosine monophosphate | dAMP |
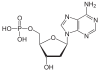
|
| Deoxyadenosine diphosphate | dADP |

|
|||
| Deoxyadenosine triphosphate | dATP |

|
|||
| Guanine | G | Deoxyguanosine | Deoxyguanosine monophosphate | dGMP |

|
| Deoxyguanosine diphosphate | dGDP |

|
|||
| Deoxyguanosine triphosphate | dGTP |
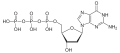
|
|||
| Cytosine | C. | Deoxycytidine | Deoxycytidine monophosphate | dCMP |

|
| Deoxycytidine diphosphate | dCDP |
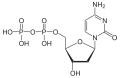
|
|||
| Deoxycytidine triphosphate | dCTP |
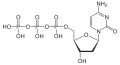
|
|||
| Thymine | T | Deoxythymidine | Deoxythymidine monophosphate | dTMP |

|
| Deoxythymidine diphosphate | dTDP |

|
|||
| Deoxythymidine triphosphate | dTTP |

|
|||
Dideoxyribose as a sugar
Artificial dideoxyribonucleoside triphosphates (ddNTPs) are used, for example, in DNA sequencing according to Sanger .
Notation of nucleotides
Letter symbols are used for the notation of the bases of nucleotides in nucleic acid sequences. In order ambiguities ( Engl. : Ambiguity ) to take into account incomplete specified nucleobases, was accepted by the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology of Ambiguity Code proposed:
| symbol | meaning | Derivation |
|---|---|---|
| G | G | G uanine |
| A. | A. | A denin |
| C. | C. | C ytosin |
| T | T | T hymin |
| U | U | U racil |
| R. | G or A | Pu r ine |
| Y | C or T | P y rimidine |
| W. | A or T | sch w oh (engl. w eak) with 2 hydrogen bonds paired |
| S. | G or C | s tark (English s trong) paired with 3 hydrogen bonds |
| M. | A or C | A m inogruppe |
| K | G or T | K etogruppe |
| H | A, C, or T (U) | not G, in the alphabet H follows G |
| B. | G, C, or T (U) | not A, in the alphabet B follows A |
| V | G, A, or C | not T (U), in the alphabet V follows U |
| D. | G, A, or T (U) | not C, in the alphabet D follows C |
| N | G, A, C or T (U) | some n your (engl. a n y) of the bases |
Note the difference between the generic symbols W, S, ... and the so-called wobble bases . There are modifications of the above symbols and additional symbols for non-standard bases and modified bases, such as the Greek letter Psi, Ψ for pseudouridine (a wobble base).
However, many of the symbols above are also used as alternatives for synthetic bases, see DNA §Synthetic bases . A selection:
- P - 2-amino - imidazo [1,2- a ] - 1,3,5-triazine - 4 (8 H ) -one and Z - 6-amino-5-nitro-2 (1 H ) -pyridone
- X - NaM, Y - 5SICS and Y '- TPT3! Other bases from this series: FEMO and MMO2
- P - 5-aza-7-deazaguanine, B - isoguanine, rS - isocytosine, dS - 1-methylcytosine and Z - 6-amino-5-nitropyridin-2-one, see Hachimoji-DNA
- xA, xT, xC, xG (analogous with prefix xx, y and yy), see xDNA
Use in biochemistry
In addition to the use of nucleotides as combined nucleic acids in DNA and RNA , nucleotides also have several functions as monomer molecules . Examples of this can mainly be found in energy supply , coenzymes and hormonal biology .
In terms of energy, nucleotides occur through adenosine triphosphate or adenosine diphosphate and adenosine monophosphate . Resistant from 3 phosphates, an adenosine group and a ribose molecule, ATP and its energetically inferior splitting products ADP and AMP can also be classified as nucleotides.
Certain coenzymes also contain nucleotide components. An example of this is the coenzyme A .
See also
- Dinucleotide
- Oligonucleotide
- Polynucleotide
- cyclic adenosine monophosphate (cAMP) and cyclic guanosine monophosphate (cGMP)
- Deoxyadenosine monoarsenate (dAMAs) see GFAJ-1 §Discussion about the incorporation of arsenic in biomolecules (questionable incorporation into DNA in Halomonas species GFAJ-1)
- Xanthosine monophosphate (XMP)
- Flavin Mononucleotide (FMN)
literature
- D. Voet, J. Voet, C. Pratt: Textbook of Biochemistry. 2nd Edition. Wiley-VCH, Weinheim 2010, ISBN 978-3-527-32667-9 , pp. 45ff, 600ff.
- Bruce Alberts among others: Textbook of Molecular Cell Biology. 4th edition. Wiley, 2012, ISBN 978-3-527-32824-6 , pp. 57-62, 80ff.
Individual evidence
- ^ Nomenclature Committee of the International Union of Biochemistry (NC-IUB): Nomenclature for Incompletely Specified Bases in Nucleic Acid Sequences. 1984, Retrieved November 16, 2019 .
- ↑ AM Sismour, S. Lutz, JH Park, MJ Lutz, PL Boyer, SH Hughes, SA Benner: PCR amplification of DNA containing non-standard base pairs by variants of reverse transcriptase from Human Immunodeficiency Virus-1. , in: Nucleic acids research . Volume 32, number 2, 2004, pp. 728-735, doi: 10.1093 / nar / gkh241 , PMID 14757837 , PMC 373358 (free full text).
- ↑ Z. Yang, D. Hutter, P. Sheng, AM Sismour and SA Benner: Artificially expanded genetic information system: a new base pair with an alternative hydrogen bonding pattern. (2006) Nucleic Acids Res. 34, pp. 6095-6101. PMC 1635279 (free full text)
-
↑ Z. Yang, AM Sismour, P. Sheng, NL Puskar, SA Benner: Enzymatic incorporation of a third nucleobase pair. , in: Nucleic acids research . Volume 35, number 13, 2007, pp. 4238-4249,
doi: 10.1093 / nar / gkm395 , PMID 17576683 , PMC 1934989 (free full text). - ↑ Yorke Zhang, Brian M. Lamb, Aaron W. Feldman, Anne Xiaozhou Zhou, Thomas Lavergne, Lingjun Li, Floyd E. Romesberg: A semisynthetic organism engineered for the stable expansion of the genetic alphabet , in: PNAS 114 (6), February 7, 2017, pp. 1317-1322; First published January 23, 2017, doi: 10.1073 / pnas.1616443114 , Ed .: Clyde A. Hutchison III, The J. Craig Venter Institute
- ↑ scinexx: Researchers breed "Frankenstein" microbe , from January 24, 2017
-
↑ Indu Negi, Preetleen Kathuria, Purshotam Sharma, Stacey D. Wetmore How do hydrophobic nucleobases differentiate from natural DNA nucleobases? Comparison of structural features and duplex properties from QM calculations and MD simulations , in: Phys. Chem. Chem. Phys. 2017, 19, pp. 16305-16374,
doi: 10.1039 / C7CP02576A - ↑ Shuichi Hoshika, Nicole A. Leal, Myong-Jung Kim, Myong-Sang Kim, Nilesh B. Karalkar, Hyo-Joong Kim, Alison M. Bates, Norman E. Watkins Jr., Holly A. Santa Lucia, Adam J. Meyer , Saurja DasGupta, Joseph A. Piccirilli, Andrew D. Ellington, John SantaLucia Jr., Millie M. Georgiadis, Steven A. Benner: Hachimoji DNA and RNA: A genetic system with eight building blocks. Science 363 (6429), February 22, 2019, pp. 884-887, doi: 10.1126 / science.aat0971 .
- ↑ Daniela Albat: DNA with eight letters , on: scinexx and Extended Code of Life , on: Wissenschaft.de (bdw online), both from February 22, 2019.
-
^ SR Lynch, H. Liu, J. Gao, ET Kool: Toward a designed, functioning genetic system with expanded-size base pairs: solution structure of the eight-base xDNA double helix. , in: Journal of the American Chemical Society . Volume 128, No. 45, November 2006, pp. 14704-14711,
doi: 10.1021 / ja065606n . PMID 17090058 . PMC 2519095 (free full text). - ^ Nucleotides - Biology. Retrieved February 13, 2020 .
- ^ Bugg, Tim .: Introduction to enzyme and coenzyme chemistry . 3rd ed. Wiley, Chichester, West Sussex 2012, ISBN 978-1-118-34899-4 .