Genetic code
As genetic code which is referred to with which the nucleotide sequence of an RNA -Einzelstrangs in the amino acid sequence of the polypeptide chain of a protein translated is. This happens in the cell after the genetic information stored in the sequence of base pairs of the DNA double strand has been rewritten into the sequence of the RNA single strand (messenger or messenger ribonucleic acid , mRNA) .
This genetic code is basically the same for all known types of living being. He assigns a triplet of three successive nucleobases of the nucleic acids - the so-called codon - to a specific proteinogenic amino acid . The translation, called translation , takes place on the ribosomes in the cytosol of a cell . According to the specification of the sequence of nucleotides of an mRNA, they form the sequence of amino acids of a peptide by assigning a specific amino acid to each codon via the anticodon of a transfer ribonucleic acid (tRNA) and connecting this to the previous one. In this way, certain predetermined information is converted into the form of a peptide chain, which then folds into a special form of a protein .
However, the more complex living beings, the higher the proportion of genetic information that is not translated into proteins seems to be. A considerable amount of non-coding DNA is transcribed into RNAs , but not translated into a peptide chain. These non-protein-coding RNA species of the transcriptome include the tRNAs and ribosomal RNAs ( rRNA ) required for translation and a number of other, mostly small, RNA forms. These serve in a variety of ways to regulate various cellular processes - such as the transcription itself, as well as the possible translation, as well as a possible DNA repair , and also special epigenetic markings of DNA sections and u. a. various functions of the immune system .

GCCThe transfer ribonucleic acids, tRNAs, contain a distinctive nucleotide triplet that distinguishes them from one another at a prominent position in a loop of the cloverleaf-like molecule. It consists of three nucleotides each, which correspond to the nucleotides of a certain codon by being complementary to these and thus forming a tripartite anticodon . Codon and anticodon are base-paired and have the same specific amino acid assigned to them. Each tRNA is loaded with the amino acid for which the codon corresponding to its anticodon stands. In this way, through the specific binding of an amino acid to a tRNA with a certain anticodon, the symbol for a certain amino acid, the codon, is translated into the genetically encoded amino acid.
Strictly speaking, the genetic code is already contained in the structure of the various tRNA types: Each tRNA molecule contains an amino acid binding site structured in such a way that only the amino acid that corresponds to its anticodon according to the genetic code is bound to it. After binding to its tRNA, an amino acid is available for the biosynthesis of proteins on the ribosome so that it can be added as the next link in the polypeptide chain - if the anticodon of the tRNA matches a codon in the given nucleotide sequence of the mRNA.
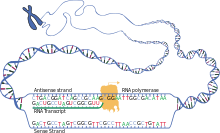
As a prerequisite for this protein synthesis, the DNA segment of a gene must first be rewritten into a ribonucleic acid (RNA) ( transcription ). In eukaryotic cells, certain parts of this hnRNA can be specifically removed ( splicing ) or changed afterwards ( RNA editing ); This preliminary pre-mRNA is then processed further to form the definitive mRNA , which is then exported from the cell nucleus. Only on the ribosomes, which can be freely present in the cytosol or are bound to the endoplasmic reticulum , are the amino acids of the tRNAs that match the codons linked to form a polypeptide using the mRNA template .
This process, with which the information of a gene is expressed in the form of a protein ( gene expression ), results from a series of steps. Here, the main processes are distinguished as (1) transcription - a section of the DNA of the genome is transcribed into RNA by RNA polymerase - and (2) post-transcriptional modification - an RNA of the transcriptome is changed - and (3) translation - an mRNA is on Ribosome translated into a polypeptide. This can be followed by a post-translational modification (4) - a polypeptide of the proteome is changed. In the course of these processes up to the provision of a functional protein, translation is the step in which the genetic information of the base triplet sequence is converted into an amino acid sequence.
The actual application of the genetic code, namely the translation of a nucleotide sequence into an amino acid based on the codon or the anticodon, takes place when an amino acid is bound to its tRNA by the respective aminoacyl-tRNA synthetase , i.e. when the amino acids are prepared for theirs possible assembly in one protein. A few base triplets do not code for an amino acid. Insofar as they have no meaning in this sense, they are also called nonsense codons ; During translation, these lead to a stop that terminates protein synthesis and are therefore also called stop codons .
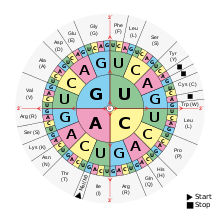
Basically, all living beings use the same genetic code. The most frequently used version is shown in the following tables . For this standard code, they show which amino acids are commonly encoded by one of the 4 3 = 64 possible codons , or which codon is translated into one of the 20 canonical amino acids . For example, the codon stands for the amino acid aspartic acid (Asp), and cysteine (Cys) is encoded by the codons and . The bases given in the table are adenine (A), guanine (G), cytosine (C) and uracil (U) of the ribonucleotides of the mRNA; in the nucleotides of the DNA, however, thymine (T) occurs instead of uracil. When a DNA segment is transcribed, an RNA polymerase uses the codogenic strand as a template for the transcript: the base- pairing DNA base sequence is rewritten into the complementary RNA base sequence when an RNA strand is built up. In this way, the genetic information stored in DNA can be accessed, which is then available in mRNA for protein biosynthesis .
GAUUGUUGC
Story of discovery
In the first half of the 1960s there was some competition among biochemists for understanding the genetic code. On May 27, 1961 at 3 a.m., the German biochemist Heinrich Matthaei achieved the decisive breakthrough in Marshall Nirenberg's laboratory with the Poly-U experiment : the decoding of the codon UUUfor the amino acid phenylalanine . This experiment is described by some geneticists as the most important of the 20th century. In 1966, five years after the first codon had been deciphered, the genetic code was completely deciphered with all 64 base triplets.
Codon
Genetic information for the structure of proteins is contained in certain sections of the base sequence of nucleic acids. Rewritten (transcribed) from DNA into RNA, it becomes available for the biosynthesis of proteins. The base sequence in the open reading frame is read from the ribosome and translated into the amino acid sequence of the synthesized peptide chain, the primary structure of a protein, according to the genetic code . The base sequence is read step by step, broken down into groups of three, and each triplet is assigned a matching tRNA loaded with a specific amino acid. The amino acid is linked to the previous one by peptide bonding. In this way the sequence segment codes for protein.
The codon is the variation pattern of a sequence of three nucleobases of the mRNA , a base triplet that can code for an amino acid . There are a total of 4 3 = 64 possible codons, of which 61 code for the 20 canonical of the proteinogenic amino acids; the remaining three are so-called stop codons for terminating translation. Under certain circumstances, these can be used to encode two additional non-canonical amino acids. This means that there are several different codings for almost all of the amino acids, most of them quite similar. Coding as a triplet is necessary insofar as a doublet coding would result in only 4 2 = 16 possible codons, so that there would not be enough possibilities for the twenty canonical or standard amino acids.
| 2nd base | |||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| U | C. | A. | G | ||||||||||||||||||||||||||||||||||
| 1st base | U |
|
|
|
|
||||||||||||||||||||||||||||||||
| C. |
|
|
|
|
|||||||||||||||||||||||||||||||||
| A. |
|
|
|
|
|||||||||||||||||||||||||||||||||
| G |
|
|
|
|
|||||||||||||||||||||||||||||||||
|
* The triplet |
||||||||||||||||||||||||||||||||||||
The codons given apply to the nucleotide sequence of an mRNA . It is read in the 5 '→ 3' direction on the ribosome and translated into the amino acid sequence of a polypeptide .
| Az | AS | AS | Codon |
|---|---|---|---|
| 1 | begin | > | AUG |
| 1 | Mead | M. | AUG |
| 1 | Trp | W. | UGG |
| 1 | Sec | U | (UGA) |
| 1 | Pyl | O | (UAG) |
| 2 | Tyr | Y | UAU UAC |
| 2 | Phe | F. | UUU UUC |
| 2 | Cys | C. | UGU UGC |
| 2 | Asn | N | AAU AAC |
| 2 | Asp | D. | GAU GAC |
| 2 | Gln | Q | CAA CAG |
| 2 | Glu | E. | GAA GAG |
| 2 | His | H | CAU CAC |
| 2 | Lys | K | AAA AAG |
| 3 | Ile | I. | AUU AUC AUA |
| 4th | Gly | G | GGU GGC GGA GGG |
| 4th | Ala | A. | GCU GCC GCA GCG |
| 4th | Val | V | GUU GUC GUA GUG |
| 4th | Thr | T | ACU ACC ACA ACG |
| 4th | Per | P | CCU CCC CCA CCG |
| 6th | Leu | L. | CUU CUC CUA CUG UUA UUG |
| 6th | Ser | S. | UCU UCC UCA UCG AGU AGC |
| 6th | Arg | R. | CGU CGC CGA CGG AGA AGG |
| 3 | stop | < | UAA UAG UGA |
The translation begins with a start codon . However, certain initiation sequences and factors are also necessary to bring about the binding of the mRNA to a ribosome and to start the process. This also includes a special initiator tRNA that carries the first amino acid. The most important start codon is the one AUGthat codes for methionine . Also, ACGand CUG- and GUG, and UUGin prokaryotic cells - serve as a start codon, but with less efficiency. The first amino acid, however, is mostly a methionine - in bacteria and in mitochondria N -formylated .
The translation ends with one of the three stop codons , also called termination codons. Initially, these codons were also given names - UAGis amber (amber-colored), UGAis opal (opal-colored), and UAAis ocher (ocher-colored) (a play on the surname of its discoverer Harris Bernstein).
While the codon is UGAmostly read as a stop , it can rarely and only under certain conditions stand for a 21st (proteinogenic) amino acid: selenocysteine (Sec). The biosynthesis and the mechanism of incorporation of selenocysteine into proteins are very different from that of all other amino acids: its insertion requires a novel translation step in which one is UGAinterpreted differently within a certain sequence environment and together with certain cofactors. This also requires a structurally unique tRNA (tRNA Sec ) intended for selenocysteine , which in vertebrates can also be loaded with two chemically related amino acids: in addition to selenocysteine, serine or phosphoserine .
Some archaea and bacteria can UAGalso translate a canonical stop codon into another (22nd) proteinogenic amino acid: pyrrolysine (Pyl). They have a special tRNA Pyl and a specific enzyme to load them (pyrrolysyl- tRNA synthetase ).
Some short DNA sequences are rarely or not at all in the genome of a species ( nullomeres ). In the case of bacteria , some of these prove to be toxic; also the codon AGA, which codes for the amino acid arginine , is avoided in bacteria (used instead CGA). There are species-specific differences in the use of codons . Differences in the use of codons do not necessarily mean differences in the frequency of amino acids used. Because for most of the amino acids there is more than a single codon, as the table above shows.
Degeneration and forgiveness
If a certain amino acid is to be coded, one can often choose from several codons with the same meaning. The genetic code is a code in which several expressions have the same meaning, i.e. the same semantic unit can be encoded by different syntactic symbols. In comparison to a coding system in which each semantic unit corresponds to a syntactic expression and vice versa, such a code is called degenerate .
It is advantageous that over 60 codons are available for the approximately 20 amino acids to be incorporated translationally. They are each represented as a combination of three nucleotides with four possible bases each , so that there are 64 combinations. Their assignment to an amino acid is such that very similar codon variations code for a specific amino acid. Due to the fault tolerance of the genetic code, two nucleotides are often enough to reliably identify an amino acid.

The base triplets coding for an amino acid usually differ in only one of the three bases; they have the minimum distance in the code space, see Hamming distance or Levenshtein distance . Mostly triplets differ in the third base, the "wobble", which is most likely to be misread in translations (see "wobble" hypothesis ). Amino acids that are often required for protein synthesis are represented by more codons than rarely used. A deeper analysis of the genetic code reveals further relationships, for example with regard to the molar volume and the hydrophobic effect (see figure).
It is also noteworthy that the base in the middle of a triplet can largely indicate the character of the assigned amino acid: So in the case of _ U _ it is hydrophobic, but hydrophilic in the case of _ A _. With _ C _ it is non-polar or polar without charge, those with charged side chains occur with _ G _ as well as with _ A _, with a negative charge only with _ A _ (see table above). Therefore radical substitutions - the exchange for amino acids of a different character - are often the result of mutations in that second position. Mutations in the third position ("wobble"), on the other hand, often preserve the respective amino acid or at least its character as a conservative substitution . Since transitions (conversion of purines or pyrimidines into one another, e.g. C → T ) occur more frequently than transversions (conversion of a purine into a pyrimidine or vice versa; this process usually requires depurination ) for mechanistic reasons , there is a further explanation for the conservative ones Properties of the code.
Contrary to previous assumptions, the first codon position is often more important than the second position, presumably because changes in the first position alone can reverse the charge (from a positively charged to a negatively charged amino acid or vice versa). However, a charge reversal can have dramatic consequences for protein function. This was overlooked in many previous studies.
The so-called degeneration of the genetic code also makes it possible to store the genetic information less sensitive to external influences. This is particularly true with regard to point mutations , both for synonymous mutations (which lead to the same amino acid) and for non- synonymous mutations which lead to amino acids with similar properties.
Obviously, early on in evolutionary history, it was helpful to reduce the susceptibility of the coding to incorrectly formed codons. The function of a protein is determined by its structure. This depends on the primary structure , the sequence of the amino acids : how many, which and in what order are linked to form a peptide chain . This information contains the base sequence as genetic information. An increased error tolerance of the coding ensures correct decoding. If an amino acid with a similar character is incorporated into the wrong one, this changes the protein function less than if it were a completely different character.
Origin of the genetic code
The use of the word “code” goes back to Erwin Schrödinger , who used the terms “hereditary code-script”, “chromosome code” and “miniature code” in a series of lectures in 1943, which he summarized in 1944 and used as the basis for his book “ What is life? ”From 1944. The exact location or carrier of this code was still unclear at the time.
It used to be believed that the genetic code came about by chance. In 1968, Francis Crick still described it as a “frozen coincidence”. However, it is the result of strict optimization in terms of fault tolerance. Errors are particularly serious for the spatial structure of a protein if the hydrophobicity of an incorrectly incorporated amino acid differs significantly from the original. In a statistical analysis, out of a million random codes, only 100 are better than the real one. If additional factors are taken into account when calculating the error tolerance, which correspond to typical patterns of mutations and reading errors, this number is even reduced to 1 in 1 million.
Universality of code
Basic principle
It is noteworthy that the genetic code is in principle the same for all living beings, so all living beings use the same “genetic language”. It is not just that genetic information is always present in the sequence of nucleic acids and is always read in triplets for the structure of proteins. With a few exceptions, a specific codon always stands for the same amino acid; the standard code reflects the common usage. It is therefore possible in genetic engineering such. B. smuggling the gene for human insulin into bacteria so that they then produce the hormone protein insulin. This common basic principle of coding, which is shared by all organisms, is known as the “universality of code”. Evolution can explain that the genetic code was formed very early in the history of life and was then passed on by all developing species. Such a generalization does not exclude that the frequency of different codewords can differ between the organisms (see Codon Usage ).
variants
In addition, there are also various variants that deviate from the standard code, in which a few codons are translated into an amino acid other than the one specified in the #standard codon table . Some of these deviations can be limited taxonomically so that special codes can be defined. In this way, over thirty variant genetic codes are now differentiated.
In eukaryotic cells, those organelles that have their own genomic system and presumably descend from symbiotic bacteria ( endosymbiont theory ) show their own variants of the genetic code. In mitochondria , ten modified forms of mitochondrial codes are known for their own DNA ( mtDNA , mitogenome syn. Chondrioma ) . These differ from the nuclear code for the genetic material in the nucleus , the nucleus genome ( karyoma ). In addition, the plastids that also occur in plant cells have their own code for their plastid DNA ( cpDNA , plastome ).
The ciliate animals (Ciliophora) also show deviations from the standard code: UAGand often also UAAcode for glutamine; this deviation is also found in some green algae . UGAalso sometimes stands for cysteine. Another variant is found in the yeast Candida , where CUGserine codes.
Furthermore, there are some variants of amino acids that can be incorporated not only by bacteria (Bacteria) and Archaea (Archaea) during translation by recoding; thus UGA, as described above, selenocysteine and UAG pyrrolysine can code, in the standard code both stop codons .
In addition, other deviations from the standard code are known, which often relate to initiation (start) or termination (stop); In mitochondria in particular, a codon (base triplet of the mRNA) is often not assigned the usual amino acid. Some examples are given in the following table:
| Occurrence | Codon | default | deviation |
|---|---|---|---|
| Mitochondria (in all organisms examined so far) | UGA | stop | Tryptophan |
| Mammalian, Drosophila and S. cerevisiae mitochondria and protozoa | AUA | Isoleucine | Methionine = start |
| Mammalian mitochondria | AGC, AGU | Serine | stop |
| Mammalian mitochondria | AG (A, G) | Arginine | stop |
| Mitochondria of Drosophila | AGA | Arginine | stop |
| Mitochondria z. B. in Saccharomyces cerevisiae | CU (U, C, A, G) | Leucine | Threonine |
| Mitochondria of higher plants | CGG | Arginine | Tryptophan |
| Some species of the genus Candida | CUG | Leucine | Serine |
| Eukarya (rare) | CUG | Leucine | begin |
| Eukarya (rare) | ACG | Threonine | begin |
| Eukarya (rare) | GUG | Valine | begin |
| Bacteria | GUG | Valine | begin |
| Bacteria (rare) | UUG | Leucine | begin |
| Bacteria (SR1 Bacteria) | UGA | stop | Glycine |
Genetic codes in DNA alphabet
DNA - sequence databases such as GenBank also be mRNA sequences in a historic conventions appropriate format in which the DNA alphabet is used, that is T instead of U stands. Examples:
- Standard code (= id )
AS = FFLLSSSSYY**CC*WLLLLPPPPHHQQRRRRIIIMTTTTNNKKSSRRVVVVAAAADDEEGGGG Starts = ---M------**--*----M---------------M---------------------------- Base1 = TTTTTTTTTTTTTTTTCCCCCCCCCCCCCCCCAAAAAAAAAAAAAAAAGGGGGGGGGGGGGGGG Base2 = TTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGG Base3 = TCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAG
id = FFLLSSSSYY**CC*WLLLLPPPPHHQQRRRRIIIMTTTTNNKKSSRRVVVVAAAADDEEGGGG
- Vertebrate Mitochondrial Code
AS = FFLLSSSSYY**CCWWLLLLPPPPHHQQRRRRIIMMTTTTNNKKSS**VVVVAAAADDEEGGGG Starts = ----------**--------------------MMMM----------**---M------------ Base1 = TTTTTTTTTTTTTTTTCCCCCCCCCCCCCCCCAAAAAAAAAAAAAAAAGGGGGGGGGGGGGGGG Base2 = TTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGG Base3 = TCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAG
id = FFLLSSSSYY**CC*WLLLLPPPPHHQQRRRRIIIMTTTTNNKKSSRRVVVVAAAADDEEGGGG
- Yeast Mitochondrial Code
AS = FFLLSSSSYY**CCWWTTTTPPPPHHQQRRRRIIMMTTTTNNKKSSRRVVVVAAAADDEEGGGG Starts = ----------**----------------------MM---------------------------- Base1 = TTTTTTTTTTTTTTTTCCCCCCCCCCCCCCCCAAAAAAAAAAAAAAAAGGGGGGGGGGGGGGGG Base2 = TTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGG Base3 = TCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAG
id = FFLLSSSSYY**CC*WLLLLPPPPHHQQRRRRIIIMTTTTNNKKSSRRVVVVAAAADDEEGGGG
- Invertebrate Mitochondrial Code
AS = FFLLSSSSYY**CCWWLLLLPPPPHHQQRRRRIIMMTTTTNNKKSSSSVVVVAAAADDEEGGGG Starts = ---M------**--------------------MMMM---------------M------------ Base1 = TTTTTTTTTTTTTTTTCCCCCCCCCCCCCCCCAAAAAAAAAAAAAAAAGGGGGGGGGGGGGGGG Base2 = TTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGG Base3 = TCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAG
id = FFLLSSSSYY**CC*WLLLLPPPPHHQQRRRRIIIMTTTTNNKKSSRRVVVVAAAADDEEGGGG
AS = FFLLSSSSYY**CC*WLLLLPPPPHHQQRRRRIIIMTTTTNNKKSSRRVVVVAAAADDEEGGGG Starts = ---M------**--*----M------------MMMM---------------M------------ Base1 = TTTTTTTTTTTTTTTTCCCCCCCCCCCCCCCCAAAAAAAAAAAAAAAAGGGGGGGGGGGGGGGG Base2 = TTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGGTTTTCCCCAAAAGGGG Base3 = TCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAGTCAG
id = FFLLSSSSYY**CC*WLLLLPPPPHHQQRRRRIIIMTTTTNNKKSSRRVVVVAAAADDEEGGGG
Note: In the first line “AS”, the amino acids are given in the single-letter code (see # Reversed codon table ), with deviations from the standard code (id) being shown in bold (or red). In the second line “Starts” shows MInitiation, *Termination; some variants differ only with regard to (alternative) start codons or stop codons . Further codes can be found in the freely accessible source.
Genetic code engineering
Generally the concept of the evolution of the genetic code from the original and ambiguous genetic code to the well-defined ("frozen") code with the repertoire of 20 (+2) canonical amino acids is accepted. However, there are different opinions and ideas about how these changes came about. Based on these, models are even proposed that predict “entry points” for the invasion of the genetic code with synthetic amino acids.
See also
literature
- Lily E. Kay: Who wrote the book of life? A history of the genetic code. Stanford University Press, Stanford, Calif. 2000
- German edition: The book of life. Who wrote the genetic code? Translated from American English by Gustav Roßler. Suhrkamp, Frankfurt am Main 2005, ISBN 3-518-29346-X .
- Rüdiger Vaas: The genetic code. Evolution and self-organized optimization, deviations and targeted change . Wissenschaftliche Verlagsgesellschaft, Stuttgart 1994, ISBN 3-8047-1383-1 .
- Lei Wang, Peter G. Schultz : The Extension of the Genetic Code . In: Angewandte Chemie. Volume 117, No. 1, 2005, pp. 34-68, doi: 10.1002 / anie.200460627 .
Web links
- The Evolution of the Genetic Code: New Findings Support the "Stereochemical Model"
- webmic.de
- Online DNA → Amino Acid Converter
- Standard code and exceptions
- Gene Cards: Human Gene Database (English)
Individual evidence
- ↑ H. Drabkin, U. RajBhandary: Initiation of protein synthesis in mammalian cells with codons other than AUG and amino acids other than methionine. In: Molecular and Cellular Biology. Volume 18, Number 9, September 1998, pp. 5140-5147; PMID 9710598 . PMC 109099 (free full text).
- ↑ LR Cruz-Vera, MA Magos-Castro, E. Zamora-Romo, G. Guarneros: Ribosome stalling and peptidyl-tRNA drop-off during translational delay at AGA codons. In: Nucleic acids research. Volume 32, Number 15, 2004, pp. 4462-4468. doi: 10.1093 / nar / gkh784 . PMID 15317870 . PMC 516057 (free full text).
- ↑ M. dos Reis, R. Savva, L. Wernisch: Solving the riddle of codon usage preferences: a test for translational selection. In: Nucleic acids research. Volume 32, Number 17, 2004, pp. 5036-5044. doi: 10.1093 / nar / gkh834 . PMID 15448185 . PMC 521650 (free full text).
- ^ U. Lagerkvist: "Two out of three": an alternative method for codon reading. In: Proceedings of the National Academy of Sciences . Volume 75, Number 4, April 1978, pp. 1759-1762. PMID 273907 . PMC 392419 (free full text).
- ↑ J. Lehmann, A. Libchaber: Degeneracy of the genetic code and stability of the base pair at the second position of the anticodon. In: RNA. Volume 14, Number 7, July 2008, pp. 1264-1269. doi: 10.1261 / rna.1029808 . PMID 18495942 . PMC 2441979 (free full text).
- ↑ Markus Fricke, Ruman Gerst, Bashar Ibrahim, Michael Niepmann, Manja Marz: Global importance of RNA secondary structures in protein coding sequences . In: Bioinformatics . August 7, 2018, doi : 10.1093 / bioinformatics / bty678 (English).
- ↑ James Dewey Watson, Tania A. Baker, Stephen P. Bell, Alexander Gann, Michael Levine, Richard Losick, et al .: Molecular Biology of the Gene . 6th edition. Pearson / Benjamin Cummings, San Francisco 2008, ISBN 978-0-8053-9592-1 , pp. 521 ff .
- ^ Erwin Schrödinger : What is life? The Physical Aspect of the Living Cell . 1944 ( whatislife.stanford.edu [PDF] Based on lectures delivered under the auspices of the Dublin Institute for Advanced Studies at Trinity College, Dublin, in February 1943).
- ^ Francis Crick : The Origin of the Genetic Code (= Journal of Molecular Biology . Volume 38 ). Elsevier, 1968, ISSN 0022-2836 , pp. 367-79 .
- ↑ Stefan Klein : All coincidence: The force that determines our life . 2015, ISBN 978-3-499-61596-2 ( books.google.de ).
- ^ CR Woese: On the Evolution of the Genetic Code. PNAS, 1965, pp. 1546-1552, PMC 300511 (free full text).
- ↑ Guenther Witzany: Crucial steps to life: From chemical reactions to code using agents . In: Biosystems . tape 140 , February 1, 2016, p. 49-57 , doi : 10.1016 / j.biosystems.2015.12.007 .
- ↑ Stephen J. Freeland, Laurence D. Hurst: The Refined Code of Life . Spectrum of Science , July 2004, p. 86-93 .
- ↑ V. Kubyshkin, CG Acevedo-Rocha, N. Budisa: On universal coding events in protein biogenesis . In: Biosystems . 2017. doi : 10.1016 / j.biosystems.2017.10.004 .
- ↑ a b c The Genetic Codes , according to NCBI, last updated: November 18, 2016; Retrieved October 25, 2017.
- ^ JH Campbell, P. O'Donoghue et al. a .: UGA is an additional glycine codon in uncultured SR1 bacteria from the human microbiota. In: Proceedings of the National Academy of Sciences . Volume 110, Number 14, April 2013, pp. 5540-5545. doi: 10.1073 / pnas.1303090110 . PMID 23509275 . PMC 3619370 (free full text).
- ↑ Nediljko Budisa: The book at the Wiley Online Library . Wiley-VCH-Verlag, Weinheim 2005, ISBN 978-3-527-31243-6 , doi : 10.1002 / 3527607188 (English).
- ↑ V. Kubyshkin, N. Budisa: Synthetic alienation of microbial organisms by using genetic code engineering: Why and how? . In: Biotechnology Journal . 12, 2017, p. 1600097. doi : 10.1002 / biot.201600097 .