Chloroplast
| Parent |
|
Organelle plastid |
| Subordinate |
|
Membrane intermembrane space Thylakoid |
| Gene Ontology |
|---|
| QuickGO |
The chloroplasts (from ancient Greek χλωρός chlōrós "green" and πλαστός plastós "shaped") are organelles of the cells of green algae and land plants that carry out photosynthesis . In higher plants , chromoplasts , leucoplasts ( amyloplasts , elioplasts ), etioplasts and gerontoplasts (collectively referred to as plastids ) can develop from photosynthetically active chloroplasts through differentiation .
Build up of chloroplasts
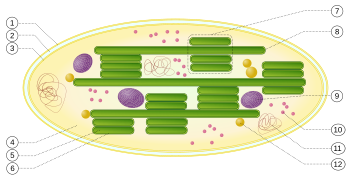
1 : outer envelope membrane
2 : intermembrane space
3 : inner envelope membrane (1 + 2 + 3: envelope)
4 : stroma (matrix)
5 : thylakoid lumen (inside the thylakoid)
6 : thylakoid membrane
7 : granum (granal lamella)
8 : thylakoid (Stromalamelle)
9 : starch body
10 : plastid ribosome ( Plastoribosom )
11 : plastid DNA ( cpDNA syn ctDNA.)
12 : Plastoglobulus (spherical structure of lipids; pl .: plastoglobules)
The thylakoid membranes are either in a stacked ( 7 ) or in unstacked ( 8 ) form.

A : The proplastid from unexposed plants consists only of an inner and an outer membrane.
B : Light induces the synthesis of chlorophyll , phospholipids and thylakoid proteins. Vesicles constrict from the inner membrane into the matrix space.
C : During the enlargement of the proplastids, individual thylakoids are formed by the fusion of vesicles. A protein in the light-harvesting complex then causes the thylakoids to stick together (stack).
D : By expanding the membrane systems, whereby the grana-thylakoid stacks are connected by (unstacked) stroma-thylakoids, the chloroplast is created
The chloroplasts of land plants have a diameter of about 4 to 8 µm . With many algae with only a single chloroplast per cell, however, this can take up a large part of the cell.
The structure of the chloroplasts is similar to that of the cyanobacteria (blue-green bacteria, formerly called blue-green algae). Chloroplasts (almost always) have their own plastid DNA ( chloroplast DNA , abbreviated cpDNA or ctDNA) together with their own ribosomes (plastid ribosomes or plastoribosomes) and are similar to mitochondria . The genome of the chloroplasts and the other plastids is also known as the plastome .
Chloroplasts are encased in two biomembranes , inside of which the stroma is located as a plasmatic phase . The stroma in turn is traversed by thylakoid membranes , descendants of the inner membrane. With the exception of many phototrophic protists , in the chloroplasts of the higher phototrophs, flat, round protuberances of these membranes are superimposed “like a roll of money” in several places - such a thylakoid stack is called granum (pl. Grana). Various pigments are embedded in the membranes of the thylakoids, especially the green pigment chlorophyll . A lot of it is found in the grana's membranes, which is why they appear intensely green in color.
The pigments can absorb light of certain wavelengths and the energy absorbed is used to produce ATP from ADP and phosphate (see phototrophy ). ATP serves as an energy carrier to build up glucose or starch from CO 2 and water.
Development of a chemiosmotic membrane potential
The biogenesis of these three membrane systems explains the fact that the membrane potential is built up by a proton gradient in chloroplasts across the thylakoid membrane (the thylakoid interior has an acidic environment), while in mitochondria the intermembrane space (area between the inner and outer membrane ) is chemiosmotic with H. + Ions is loaded. Similarly, in chloroplasts, ATP synthase (alias F o F 1 - ATPase ) is an enzyme embedded in the thylakoid membrane (CF 1 part protrudes into the stroma), in mitochondria it is part of the inner membrane (F 1 part facing the matrix). In both systems, ATP is released to the matrix / stroma. In exchange for ADP, it can get into the cytosol of the cell.
Plastoskeleton
Today we know that bacteria also have a cytoskeleton whose proteins show evolutionary relationship to those of the eukaryotic cytoskeleton. Experiments on moss Physcomitrella patens (including with knockout mosses ) is known that the FtsZ proteins, the tubulin - homologues , not only the division of chloroplasts cause, but also a complex network can form in the chloroplasts. Since these networks are strongly reminiscent of the cytoskeleton, Ralf Reski coined the term “plastoskeleton” in 2000 for these complex structures and postulated that they fulfill similar functions in the plastids as the cytoskeleton does for the entire cell .
Chloroplast DNA
Chloroplast DNA was detected for the first time in 1962, and a plastome was sequenced for the first time in 1986. Since then, hundreds of chloroplast DNAs from different species have been sequenced. Most of the time, however, these are chloroplastida , i. H. Land plants or green algae. Glaucophytes (Glaucophyta), red algae (Rhodophyta syn. Rhodophyceae) and other groups of algae are strongly underrepresented. The DNA of the chloroplasts - at least in the case of the chloroplastida (green algae and land plants) - is usually structured like most bacteria in a ring. The chloroplast genome (plastome) has portions as opposing copies ( English inverted repeats are present), similar to the genome of cyanobacteria . In mitochondria and plastids such as chloroplasts, as in bacteria, the DNA is usually condensed into nuceloids (nuclear equivalents). This is ensured by the so-called histone-like proteins (HLPs, after English h iStone l ike p roteins ). These are homologous to one another , but only functionally similar ( analogous ) to the real histones in the nucleus of the eukaryotic cells ( eucytes ). The protein- coding genes are transcribed into messenger RNA , which is used as a template for protein synthesis ( translation ) on one's own ribosomes (plastoribosomes). These plastid ribosomes are similar in structure and structure to those of bacteria and mitochondria, but are significantly smaller and simpler than those of the euzytes surrounding them.
Origin of chloroplasts - endosymbiotic theory
The German botanist Andreas Franz Wilhelm Schimper had already established in 1883 that chloroplasts are very similar to cyanobacteria and thus already indicated a symbiotic origin of chloroplasts, so that he can be regarded as a pioneer of the later formulated endosymbiont theory . The Russian biologist Konstantin Sergejewitsch Mereschkowski took up this idea in 1905 and Ivan Wallin in 1922 and made it more concrete.

For a long time it was unknown how chloroplasts divide and change shape. Since it was not possible to cultivate isolated mitochondria or chloroplasts in vitro (in the laboratory on nutrient media), the theory could not be established until the early 1970s ( Lynn Margulis ), when DNA was detected in both types of organelle. The independent “ multiplication ” of the chloroplasts, similar to bacteria, without a structural coupling to the cell division of the surrounding eukaryotic cell, is another argument for the endosymbiont theory.
Primary endosymbiosis

A eukaryote with mitochondria once enclosed a cyanobacterium. From this then chloroplasts developed in the course of evolution, so that this event formed the starting point of a lineage of cells with both organelles. Note that the endosymbiont of the cyanobacterium already had a double membrane - the membrane of the phagosomal vacuole ( food vacuole ) was lost in this case.
The exact examination of the chloroplasts and their "host cells" has shown that this process has taken place in several stages (nested). At the beginning, about 600 million to 2 billion years ago, a free-living photosynthetic cyanobacterium entered an early eukaryotic cell (euzyte), either as food ( phagocytosis ) or as an internal parasite ( endoparasite ). However, it was able to escape the phagocytic ( phagosomal ) vacuole in which it was initially, without being dissolved by the euzyte , and became a permanent resident of this "host cell".
Endosymbiotic gene transfer
In the course of the evolution and integration of the endosymbiont, various adjustments were made. This includes, among other things, the adaptation of the chloroplast genome. The size of the genome decreased from approximately 3.5 million bases to 120–160 thousand. This corresponds to a reduction of often more than 1500 genes in cyanobacteria to around 60–100 genes in chloroplasts. The shrinking of the genome has been accompanied by the loss of genetic information and transfer into the cell nucleus , a process known as ' endosymbiotic gene transfer ' (EGT).
Import of proteins into the chloroplast
At the same time, a complex machinery for importing proteins from the cytosol into the chloroplasts developed. So you can find about 2000 proteins in the chloroplast despite the only 100 remaining genes. These leftover protein-coding genes can be roughly divided into two categories: maintenance of the genetic apparatus ( DNA polymerase , tRNAs and rRNAs ) and maintenance of photosynthetic capacity ( photosystem components and other proteins). It has not yet been fully clarified how the expression between the nucleus and the chloroplasts is synchronized . This is necessary because in all protein complexes in the chloroplast, plastid and nuclear-coded products are combined.
DNA-less chloroplasts
In 2014 a plastid without a genome was even found in the non-photosynthetically active green alga Polytomella ( Chlamydomonadales , syn. Volvocales). This shows that chloroplasts can lose their entire genome through endosymbiotic gene transfer. The situation is thus analogous to that of mitochondria , where a loss of the original function is also accompanied by a strong reduction or complete loss of the genome (see hydrogenosome and mitosome ).
The first primary endosymbiosis (main line)
It has long been debated whether the resulting primary chloroplasts originated from a single endosymbiotic event or from several independent events in different eukaryotic lineages. It is now widely believed that virtually all organisms with primary chloroplasts have a single common ancestor. The time taken cyanobacterium was apparently near the present species Gloeomargarita lithophora , this is basal in the family tree of cyanobacteria close to the genus Synechococcus . The alga Cyanophora , a glaucophyte , is one of the most primitive organisms that contain a chloroplast.
Another primary endosymbiosis ( Paulinella )
The exception is the aforementioned amoeboid Paulinella chromatophora ( Euglyphida , see below). This apparently comes from an ancestor who, independently and much later - about 90 to 500 million years ago - took in a cyanobacterium of the genus Prochlorococcus (or Synechococcus ).
Lineage and basic types of chloroplasts
Overall, all primary chloroplasts belong to one of the following four lineages (the first three with a common origin):
- the glaucophyte line:
the unicellular algae of the Glaucophyta (syn. Glaucocystaceae) have plastids that are in many respects still very similar to the cyanobacterium and are therefore often referred to as " cyanelles " or "cyanoplasts", sometimes also as "muroplasts". - the rhodophyte line:
red algae (wiss. Rhodophyta) have plastids called “ rhodoplasts ”, which still carry the antenna structure ( phycobilisomes ) of the cyanobacteria. - the chloroplastid line:
The plastids of the viridiplantae (syn. chloroplastida, green algae and higher plants ) represent the most developed plastids and carry a large variety of antenna complexes. The green plastids of algae and higher plants are called chloroplasts. - of the Paulinella line:
The plastids of the amoeboid Paulinella chromatophora ( Euglyphida ) are called chromatophores .
Secondary and other endosymbioses
Although all chloroplasts, with one exception, can probably be traced back to a single such primary endosymbiosis event, chloroplasts are still found today in an extremely wide range of organisms, some of which are not even directly related to one another. This is explained as a consequence of many secondary and even tertiary endosymbiotic events in which photosynthetically active algae (including their chloroplasts) were ingested instead of cyanobacteria. These originally eukaryotic complex plastids or chloroplasts are called secondary plastids (chloroplasts).

One eukaryotic alga is incorporated by another eukaryote and thus becomes a chloroplast with three or four membranes.
Number of membranes and nucleomorph
While primary chloroplasts only have a double membrane that is derived from their cyanobacterial ancestor, secondary chloroplasts have additional membranes outside of these two original ones. This is interpreted as a consequence of the secondary endosymbiotic events. As a result of the mostly extensive degradation of the incorporated algae, often only their (primary) chloroplast and sometimes their cell membrane and / or even a remnant of their cell nucleus , called nucleomorph , remained. The resulting complex (secondary) chloroplasts can therefore have three or four membranes: the two cyanobacterial membranes, sometimes the cell membrane of the incorporated algae and the phagosome vacuole from the host's cell membrane.
The complex (secondary and tertiary) chloroplasts were often transformed in many ways. In other cases as well - such as the apicomplexa (see below) - such additional functions make even photosynthetically no longer active plastids indispensable for the cell.
Apicomplexa
The apicomplexa are a subgroup of the alveolata. The malaria parasite Plasmodium belongs to the Apicomplexa . Many apicomplexes have a red algae-derived chloroplast called an apicoplast , which they inherited from their ancestors. The apicoplasts have lost all photosynthetic functions and do not contain photosynthetic pigments or true thylakoids.
The fact that Apicomplexa still kept their non-photosynthetic chloroplasts testifies that the chloroplasts perform other important functions in addition to photosynthesis, as mentioned above - plant chloroplasts supply plant cells with many important substances in addition to sugar, and apicoplasts do not differ in this - they synthesize Fatty acids , isopentenyl pyrophosphate and iron-sulfur clusters . In addition, they perform part of the heme synthesis ( English heme pathway ).
Dinoflagellates
In some cases, existing chloroplasts were replaced by new (complex) ones, in other cases new ones were added to existing plastids if the tasks differed greatly. In the case of dinoflagellates , the chloroplasts are mostly derived from red algae and are of the peridinin type, characterized by the carotenoid pigment peridinin together with chlorophyll a and chlorophyll c 2 . Peridinin is not found in any other group of chloroplasts and has three (occasionally only two) membranes, i.e. H. they have lost the original cell membrane of the red algae endosymbiont.
Some dinoflagellates such as Kryptoperidinium and Durinskia (both Peridiniaceae , also English dinotoms ) have a tertiary chloroplast derived from diatoms (syn. Diatoms, Heterokontophyta ). These chloroplasts are surrounded by up to five membranes, depending on whether the entire diatom endosymbiont is seen as the chloroplast or only the red alga it contains is counted as the chloroplast. The diatom endosymbiont is relatively little reduced - it still contains its original mitochondria and has an endoplasmic reticulum , eukaryotic ribosomes , a cell nucleus and of course the secondary chloroplasts derived from red algae - practically a complete cell - everything inside the host. The original three-membrane peridinin chloroplast was either lost or remodeled into an eye-spot , as is believed to be the case with Durinskia .
Kleptoplastidy
Despite the reduction of the endosymbiont genome through gene transfer to the host, including the reduction of nucleomorphs (up to the point of disappearance) and of membrane envelopes in complex plastids, there is still a symbiosis as long as the endosymbiont remains capable of reproduction (which, however, in extreme cases - complete loss of the organelle genome - can end in complete assimilation). In contrast to this, kleptoplastidy describes the 'robbery' of chloroplasts, i. H. the removal of the shell and core of the recorded phototrophs (`green` eucytes), so that only the chloroplasts remain, but are no longer capable of reproduction. These so-called kleptoplasts then have to be replaced again and again by new ones. It even happens that such predators become victims again themselves. Kleptoplastidy has been observed in certain dinoflagellates , ciliates and some marine snails , but some of the genes from the cell nuclei of the food are transferred to the snails in members of the Elysia genus , which is why the chloroplasts can be supplied with proteins that are essential for them. In the case of the green hydra , however, endosymbiosis or an intermediate form is assumed.
See also
- Chemiosmotic coupling
- photosynthesis
- Calvin cycle
- C3 plant , C4 plant (with “dimorphogenic” chloroplasts) and CAM plant
- Endosymbiont theory (on the origin of chloroplasts)
- Complex plastids
- Eye spot (especially in Chlamydomonas ) and ocelloid
- Zooxanthellae
- Kleptoplastid
- Cell compartment
literature
- D. von Wettstein (1959): The effect of genetic factors on the submicroscopic structures of the chloroplast , J. Ultrastruct. Res. 3, pp. 235-239.
Web links
- Structure of chloroplasts - diagram
- Chloroplast and photosynthesis / energy production in plants - graphics / animation
- Botany online (Uni Hamburg): cells and tissues - plant cells - section "chloroplasts"
- Chloroplasts smuggle genes across species
- Wilfried Probst: Early Evolution and Symbiosis , Europa-Universität Flensburg, Institute for Biology and Science Education and its Didactics: §Plastiden, accessed on April 19, 2019
Individual evidence
- ↑ C. Cleveland (ed.); C. Michael Hogan, S. Draggan: DNA ( December 25, 2012 memento on the Internet Archive ), The Encyclopedia of Earth, National Council for Science and the Environment ( December 25, 2012 memento on the Internet Archive ). Washington DC, July 18, 2012 (via WebArchive)
- ^ The Oxford Dictionary of Abbreviations 1998.
- ↑ Strepp, R. et al. (1998): Plant nuclear gene knockout reveals a role in plastid division for the homolog of the bacterial cell division protein FtsZ, an ancestral tubulin . In: Proc. Natl. Acad. Sci. USA 95 (8); 4368-4373; PMID 9539743 , PMC 22495 (free full text).
- ^ Reski, R. (2002): Rings and networks: the amazing complexity of FtsZ in chloroplasts . In: Trends in Plant Science 7 (3); 103-105; PMID 11906832 ; doi: 10.1016 / S1360-1385 (02) 02232-X .
- ↑ Kiessling, J. et al. (2000): Visualization of a cytoskeleton-like FtsZ network in chloroplasts . In: Journal of Cell Biology 151 (4); 945-950; PMID 11076976 , PMC 2169431 (free full text).
- ^ A b Leighton Then: Bioscience — Explained: Green DNA - Simple isolation, restriction and electrophoresis of chloroplast DNA . BIOSCIENCE EXPLAINED, Science and Plants for Schools, Homerton College, Cambridge 2002. Archived from the original on September 17, 2012. (via WebArchive)
- ↑ Chloroplasts and Other Plastids . University of Hamburg. Archived from the original on September 25, 2012. Retrieved March 16, 2019. (via WebArchive)
- ↑ Anna Stina Sandelius, Henrik Aronsson: The chloroplast: Interactions with the Environment . Springer, 2009, ISBN 978-3-540-68696-5 , p. 18.
- ↑ Andreas Franz Wilhelm Schimper : About the development of the chlorophyll grains and color bodies Archived from the original on October 19, 2013. In: Bot. Newspaper . 41, 1883, pp. 105-114, 121-131, 137-146, 153-162. (via WebArchive)
- ↑ Constantin S. Mereschkowsky: About the nature and origin of the chromatophores in the plant kingdom . In: Biological Centralblatt . 25, September 15, 1905, pp. 593-604. (via WebArchive)
- ↑ Ivan E. Wallin: On the nature of mitochondria. I. Observations on mitochondria staining methods applied to bacteria. II. Reactions of bacteria to chemical treatment . In: American Journal of Anatomy . 30, No. 2, 1922, pp. 203-229. doi : 10.1002 / aja.1000300203 .
- ↑ Ivan E. Wallin: On the nature of mitochondria. III. The demonstration of mitochondria by bacteriological methods. IV. A comparative study of the morphogenesis of root-nodule bacteria and chloroplasts . In: American Journal of Anatomy . 30, No. 4, 1922, pp. 451-471. doi : 10.1002 / aja.1000300404 .
- ↑ Lynn Sagan: On the Origin of Mitosing Cells. In: J. Theoretical Biology. Vol. 14, No. 3, 1967, pp. 255-274. PMID 11541392 doi: 10.1016 / 0022-5193 (67) 90079-3
- ↑ Bernhard Kegel : The rulers of the world: How microbes determine our life. DuMont, Cologne 2015, ISBN 978-3-8321-9773-5 .
- ↑ a b Biology, 8th Edition, Campbell & Reece . Benjamin Cummings (Pearson Education), 2009, ISBN 978-0-321-54325-7 , p. 516.
- ↑ John M. Archibald: The Puzzle of Plastid Evolution . In: Current Biology . 19, No. 2, 2009, pp. R81-8. doi : 10.1016 / j.cub.2008.11.067 . PMID 19174147 .
- ↑ a b c d e f g h Patrick J. Keeling: Diversity and evolutionary history of plastids and their hosts . In: American Journal of Botany . 91, No. 10, 2004, pp. 1481-1493. doi : 10.3732 / ajb.91.10.1481 . PMID 21652304 .
- ^ A b Geoffrey I. McFadden, Giel G. Van Dooren: Evolution: Red Algal Genome Affirms a Common Origin of All Plastids . In: Current Biology . 14, No. 13, 2004, pp. R514-6. doi : 10.1016 / j.cub.2004.06.041 . PMID 15242632 .
- ↑ a b c Patricia Sánchez-Baracaldo, John A. Raven, Davide Pisani, Andrew H. Knoll: Early photosynthetic eukaryotes inhabited low-salinity habitats . In: Proceedings of the National Academy of Sciences . 114, No. 37, September 12, 2017, pp. E7737 – E7745. doi : 10.1073 / pnas.1620089114 .
- ^ Ron Milo, Rob Philips: Cell Biology by the Numbers: How large are chloroplasts? . In: book.bionumbers.org . Retrieved March 16, 2019.
- ↑ a b Anna Stina Sandelius, Henrik Aronsson (eds.); E. Kim, John M. Archibald ;: Diversity and Evolution of Plastids and Their Genomes . In: The Chloroplast (= Plant Cell Monographs), Volume 13 2009, ISBN 978-3-540-68692-7 , pp. 1-39, doi : 10.1007 / 978-3-540-68696-5_1 .
- ↑ William Martin, Tamas Rujan, Erik Richly, Andrea Hansen, Sabine Cornelsen, Thomas Lins, Dario Leister, Bettina Stoebe, Masami Hasegawa, David Penny: Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus . In: Proceedings of the National Academy of Sciences . 99, No. 19, 2002, pp. 12246-12251. bibcode : 2002PNAS ... 9912246M . doi : 10.1073 / pnas.182432999 . PMID 12218172 . PMC 129430 (free full text).
- ↑ a b M. T. Clegg, BS Gaut, GH Learn Jr, BR Morton: Rates and Patterns of Chloroplast DNA Evolution . In: Proceedings of the National Academy of Sciences . 91, No. 15, 1994, pp. 6795-6801. bibcode : 1994PNAS ... 91.6795C . doi : 10.1073 / pnas.91.15.6795 . PMID 8041699 . PMC 44285 (free full text).
- ↑ Chun Y. Huang, Michael A. Ayliffe, Jeremy N. Timmis: Direct measurement of the transfer rate of chloroplast DNA into the nucleus . In: Nature . 422, No. 6927, 2003, pp. 72-6. bibcode : 2003Natur.422 ... 72H . doi : 10.1038 / nature01435 . PMID 12594458 .
- ↑ David Roy Smith, Robert W. Lee: A Plastid without a Genome: Evidence from the Nonphotosynthetic Green Algal Genus Polytomella . In: Plant Physiology . 164, No. 4, April 1, 2014, pp. 1812-1819. doi : 10.1104 / pp.113.233718 . PMID 24563281 . PMC 3982744 (free full text).
- ^ Rafael I. Ponce-Toledo, Philippe Deschamps, Purificación López-García, Yvan Zivanovic, Karim Benzerara, David Moreira: An Early-Branching Freshwater Cyanobacterium at the Origin of Plastids . In: Current Biology . 27, No. 3, 2017, pp. 386–391. doi : 10.1016 / j.cub.2016.11.056 .
- ↑ Jan de Vries, John M. Archibald: Endosymbiosis: Did Plastids Evolve from a Freshwater Cyanobacterium? . In: Current Biology . 27, No. 3, 2017, pp. R103-R105. doi : 10.1016 / j.cub.2016.12.006 .
- ^ A b Purificación López-García, Laura Eme, David Moreira: Symbiosis in eukaryotic evolution . In: Journal of Theoretical Biology . 434, 2017, pp. 20–33. doi : 10.1016 / j.jtbi.2017.02.031 .
- ^ Geoffrey I. McFadden: Chloroplast Origin and Integration . In: Plant Physiology . 125, No. 1, 2001, pp. 50-3. doi : 10.1104 / pp.125.1.50 . PMID 11154294 . PMC 1539323 (free full text).
- ^ Luis Delaye, Cecilio Valadez-Cano, Bernardo Pérez-Zamorano: How Really Ancient Is Paulinella Chromatophora? . In: PLoS Currents . 2016. doi : 10.1371 / currents.tol.e68a099364bb1a1e129a17b4e06b0c6b .
- ^ W. Probst: On the way to chloroplast formation. European University of Flensburg.
- ↑ S. Ball, C. Colleoni, U. Cenci, JN Raj, C. Tirtiaux: The evolution of glycogen and starch metabolism in eukaryotes gives molecular clues to understand the establishment of plastid endosymbiosis . In: Journal of Experimental Botany . 62, No. 6, 2011, pp. 1775-801. doi : 10.1093 / jxb / erq411 . PMID 21220783 .
- ↑ a b Robert R. Wise (Ed.); J. Kenneth Hoober: The structure and function of plastids . Springer, Dordrecht 2006, ISBN 978-1-4020-4061-0 , pp. 3-21.
- ↑ more precisely: Chlorophyta and Streptophyta / Charophyta , the latter with the land plants , scientifically Embryophyta
- ↑ Jacques Joyard, Maryse A. Block, Roland Douce: Molecular aspects of plastid envelope biochemistry . In: Eur. J. Biochem. . 199, No. 3, 1991, pp. 489-509. doi : 10.1111 / j.1432-1033.1991.tb16148.x . PMID 1868841 .
- ↑ Balbir K. Chaal, Beverley R. Green: Protein import pathways in 'complex' chloroplasts derived from secondary endosymbiosis involving a red algal ancestor . In: Plant Molecular Biology . 57, No. 3, February 2005, pp. 333-342. doi : 10.1007 / s11103-004-7848-y . PMID 15830125 .
- ↑ a b c d e f g Patrick J. Keeling: The endosymbiotic origin, diversification and fate of plastids . In: Philosophical Transactions of the Royal Society B: Biological Sciences . 365, No. 1541, 2010, pp. 729-748. doi : 10.1098 / rstb.2009.0103 . PMID 20124341 . PMC 2817223 (free full text).
- ↑ a b Sethu C. Nair, Boris Striepen: What Do Human Parasites Do with a Chloroplast Anyway? . In: PLoS Biology . 9, No. 8, 2011, p. E1001137. doi : 10.1371 / journal.pbio.1001137 . PMID 21912515 . PMC 3166169 (free full text).
- ^ A b Jeremiah D. Hackett, Donald M. Anderson, Deana L. Erdner, Debashish Bhattacharya: Dinoflagellates: A remarkable evolutionary experiment . In: American Journal of Botany . 91, No. 10, 2004, pp. 1523-34. doi : 10.3732 / ajb.91.10.1523 . PMID 21652307 .
- ↑ Jacques Joyard, Maryse A. Block, Roland Douce: Molecular aspects of plastid envelope biochemistry . In: Eur. J. Biochem. . 199, No. 3, 1991, pp. 489-509. doi : 10.1111 / j.1432-1033.1991.tb16148.x . PMID 1868841 .
- ↑ Chloroplast . In: Encyclopedia of Science . Retrieved March 20, 2019.
- ↑ Eberhard Schnepf, Malte Elbrächter: Dinophyte chloroplasts and phylogeny - A review . In: Grana . 38, No. 2-3, 1999, pp. 81-97. doi : 10.1080 / 00173139908559217 .
- ↑ E. Hehenberger, B. Imanian, F. Burki, PJ Keeling: Evidence for the retention of two evolutionary distinct plastids in dinoflagellates with diatom endosymbionts. In: Genome Biology and Evolution (2014), 6 (9), pp. 2321-2334. doi: 10.1093 / gbe / evu182
- ^ Alf Skovgaard: Role of chloroplast retention in a marine dinoflagellate . In: Aquatic Microbial Ecology . 15, 1998, pp. 293-301. doi : 10.3354 / ame015293 .
- ↑ Richard G. Dorrell, Christopher J. Howe: Integration of plastids with their hosts: Lessons learned from dinoflagellates . In: Proceedings of the National Academy of Sciences . 112, No. 33, August 18, 2015, ISSN 0027-8424 , pp. 10247-10254. bibcode : 2015PNAS..11210247D . doi : 10.1073 / pnas.1421380112 . PMID 25995366 . PMC 4547248 (free full text).
- ^ Matthew D. Johnson, David Oldach, F. Delwiche Charles, Diane K. Stoecker: Retention of transcriptionally active cryptophyte nuclei by the ciliate Myrionecta rubra . In: Nature . 445, No. 7126, January 2007, pp. 426-428. doi : 10.1038 / nature05496 . PMID 17251979 .
- ↑ Aditee Mitra: Marine Biology - The Best of Two Worlds , Spectrum of Science, April 2019, pp. 54–60
- ↑ Rumpho ME, Worful JM, Lee J, Kannan K, Tyler MS, Bhattacharya D, Moustafa A, Manhart JR: Horizontal gene transfer of the algal nuclear gene psbO to the photosynthetic sea slug Elysia chlorotica . In: Proceedings of the National Academy of Sciences of the United States of America . 105, No. 46, Nov 2008, pp. 17867-17871. bibcode : 2008PNAS..10517867R . doi : 10.1073 / pnas.0804968105 . PMID 19004808 . PMC 2584685 (free full text).


