Denaturation (biochemistry)

Denaturation denotes a structural change in biomolecules such as proteins or deoxyribonucleic acid (DNA), which in most cases is associated with a loss of the biological function of these molecules, although their primary structure remains unchanged. Denaturation can be due to physical or chemical influences.
principle

External influences can cause structural changes in biopolymers such as proteins or nucleic acids according to the principle of the least bit of compulsion . When a protein is denatured, the secondary and tertiary structure (and thus possibly also the quaternary structure ) changes without changing the sequence of the amino acids , its primary structure . However, the protein loses its original folding shape , which is also known as the native configuration or conformation of the polypeptide chain. With the loss of the fold, the function of the protein is also lost, with the exception of the intrinsically unstructured proteins . The process of denaturation can be irreversible (irreversible) or reversible (reversible), depending on whether the native form also represents the energetically most favorable state. The reverse process of denaturation is also called renaturation . Since many proteins can be renatured and their function can be restored, it was postulated that protein folding occurs during protein biosynthesis because the native form usually represents the most energetically favorable state ( Anfinsen dogma ).
Such reversible changes in the molecular structure occur, for example, in the case of heat denaturation of DNA when it is broken into single strands by heating - for example for a polymerase chain reaction (PCR) - and then cooled again. Double-stranded DNA and RNA are held together during base pairing by hydrogen bonds , which are reversibly broken when denatured. On the other hand, after an irreversible change in the molecular structure, the original spatial structure of the molecule cannot be restored. A breakfast egg, for example, experiences such changes when it is cooked, which cannot be converted to its previous state by cooling down, once it has become a "hard egg", nor by further cooking. The denaturation of proteins usually leads to the molecule being inactivated, which means that it can hardly or no longer fulfill its biological function . Denaturation is therefore also used for disinfection .
All denaturation processes have in common that covalent bonds are not split (except for the disulfide bridges in proteins) and thus the primary structure is retained. The chain structure and thus the sequence of the building blocks as the primary structure is retained. However, through the supply of energy, individual building blocks, nucleotides and amino acids or even the entire molecular chain are made to vibrate so much that other binding forces ( ionic , polar and van der Waals interactions , hydrogen bonds, hydrophobic effects ) between different areas of the molecular chain are canceled out and such ties are broken. Disulfide bridges in proteins are usually split by reduction with sulfhydryls. When proteins are denatured, a decrease in solubility is often observed, sometimes accompanied by the formation of protein aggregates . Denaturation also increases the sensitivity to degradation by proteases .
Conformational changes in biopolymers can be measured using FTIR , dual polarization interferometry , circular dichroism , QCM-D , NMR spectroscopy, and multiparametric surface plasmon resonance spectroscopy .
Denaturation due to physical influences
The most common denaturations under physical influence are heat denaturation and radiation denaturation. In physical terms, denaturation can also be caused by high pressure, vigorous stirring, shaking, by the action of ultrasound and by interface absorption.
Heat denaturation
Heat or heat denaturation is a type of denaturation in which a change in the molecular structure is brought about by increasing the temperature. Usually, no covalent chemical bonds are broken or formed by the action of heat , so the primary structure remains unchanged. Instead, hydrogen bonds are broken or newly formed, which are usually bonds between chain sections, which often results in a change in the tertiary structure of enzymes and other proteins. This usually results in a loss of biological activity and a decrease in solubility. The latter is then noticeable as “flocculation” or “coagulation”. Since in the protein folding and hydrophobic effects play a role, the denaturation is also generated by the decrease of the hydrophobic effect with increasing temperature. A heat denaturation (like other denaturations) can be reversible if the structural changes are not yet too profound, but it is often irreversible (irreversible). However, a reversal is possible under laboratory conditions with the help of centrifuges and the addition of urea . Due to the shear forces that occur , chicken eggs can be partially “decooked”, for example. The temperature at which the denaturation of the proteins begins is very different depending on the structure and organism. The enzymes of hyper- thermophilic archaea have to withstand temperatures well above 80 ° C. The denaturation by heating is increased in a slightly acidic solution. The thermal denaturation of proteins usually takes place in a relatively narrow temperature range, which is why a cooperative unfolding during the denaturation was assumed, i. H. one development favors others.
By autoclaving be pathogens to objects by means of denaturing vital biopolymers inactivated. When autoclaving, a temperature well above 100 ° C with increased pressure must be maintained for a specified time in order to sterilize safely .
Nucleic acids denature within a very narrow temperature range, also known as the “ melting point ”, which is usually above 80 ° C. The denaturation is reversible. When the nucleic acids cool down, the single strands reassemble. This process is used in molecular biology to carry out PCR in order to replicate certain genes from an organism in vitro: extracted DNA is melted in a reaction vessel at high temperatures of around 95 ° C (denaturation). The temperature is then lowered again to a certain temperature. This annealing temperature depends on the primers and is usually 2–3 ° C below their respective melting point (mostly 50 to 65 ° C). The primers contained in the solution attach to the DNA single strands (annealing or primer hybridization). The strands are then completed again (elongation) at a temperature of 68 to 72 ° C. with the aid of a Taq polymerase . The cycle of denaturation, annealing and elongation starts over. About 25 to 50 cycles are performed. The reversible denaturation of DNA is used up to 50 times to replicate a sought-after gene in an organism. In the case of denaturation, the extinction of DNA increases by about 40% at a wavelength of 260 nm.
Denaturation by pressure
Since the reaction volumes in protein folding are very small, you usually have to use a pressure of several 1000 bar to unfold proteins. Nevertheless, the high pressure treatment of food is becoming more and more important in practice. For this purpose, the food, mostly wrapped in foil, is placed in a printing medium, such as B. water, and the pressure is applied to this medium. In this "non-thermal process", a high-pressure pasteurization , undesired microorganisms and enzymes are inactivated and the food is preserved. Loss of quality, such as when using heat, is avoided.
In general, pressure affects the tertiary and quaternary structure of proteins, while the secondary structure can hardly be changed.
Denaturation by radiation
Infrared , microwaves or other long-wave radiation have a denaturing effect due to the temperature increase caused. In the case of ionizing radiation ( ultraviolet , gamma and X-rays ), additional covalent bonds, e.g. B. be broken down by nucleic acids and thus lead to DNA damage such as chain breaks (depolymerization). In addition, as a result of the bond cleavage, new covalent bonds (for example dimerization in nucleic acids) can also arise.
Denaturation due to chemical influences
The causes of protein denaturation can be, for example, chemical substances such as some acids , bases , salts (e.g. guanidinium salts ), detergents or urea . Protein structures can also be influenced by heavy metals , as the ions form complex structures with the amino acid residues and thus change the biologically active structure of the protein.
Acid and base denaturation
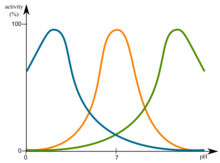
Depending on the pH value of the natural environment of the protein, proteins have a pH optimum. This optimum can be in the acidic pH range, as for example with lysosomal proteins. In other cases, however, this can also be basic. A protein is most stable in the area of the optimum pH and therefore does not denature.
The acid denaturation, for example with 40% (V / V) acetic acid, leads to charge shifts between the molecules and ultimately to a refolding of the protein into the energetically most favorable state under the respective conditions. The acid releases protons (H + ) and thus causes the charge change in the protein structure, so that the hydrogen bonds are partially destroyed and the same positive charges repel each other. In addition, the acid releases protons (H + ) to the carboxylate group (COO - ) of the amino acids aspartate and glutamate, so that carboxy groups –COOH are created and their previous negative charges disappear. This means that ionic interactions between the carboxy group and the positive charges in the protein are no longer possible.
Corresponding to bases cause, they also change the composition of the ions on the pH-value , but are amino groups of lysine or arginine deprotonated less positive charges occur thereby in the protein that could interact with negatively charged groups. In addition, carboxylic acid groups are deprotonated to carboxylates, whereby hydrogen bonds can be destroyed and more negative charges occur in the protein, which repel each other.
In the case of acid or alkali denaturation, hydrolysis of the protein can occur at the same time .
With regard to pH, the isoelectric point (pI) of a protein is also important. At this point a protein in the net charge is uncharged and therefore quickly precipitates out of solution. The protein is therefore very sensitive at the pI.
Denaturation by chaotropes
Salts and other chaotropes also have an influence on hydrophobic effects and can therefore cause denaturation, although depending on the substance, the influence can also go in the direction of renaturation. In relation to the precipitation, one speaks of “salting in” and “salting out”. The relative influence of the anions and cations that form salts is described by the “ Hofmeister series ”.
DNA is represented by formamide (70% V / V), dimethylformamide , guanidinium salts (6 M), sodium salicylate , sulfoxide , dimethyl sulfoxide (DMSO, 60% V / V), sodium hydroxide (1 M ), various alcohols , propylene glycol and urea (6 M ) denatured, usually in combination with heat. The melting temperature of the double-stranded DNA is lowered.
Denaturation by detergents
Some surfactants lead to denaturation. This is because they attach themselves all over the protein, thereby linearizing it. Ionic detergents denature the most, e.g. B. a one percent solution of sodium lauryl sulfate (SDS). Most proteins are already denatured at room temperature. In addition, membrane lipids are also released from the cell membranes , as micelles form from the surfactant and the membrane above a certain concentration . The SDS denaturation is z. B. used in sample preparation for SDS-PAGE .
Denaturation by ethanol
Corresponding to the acid denaturation, ethanol or other water-soluble organic solvents can disrupt the hydrogen bonds and hydrophobic interactions required in biopolymers to maintain the structure by interfering as polar organic solvents. 50 to 70 percent ethanol denatures most proteins. Since the membrane proteins also lose their function due to the dissolution of the membrane lipids and the denaturation of the spatial structure and the cells in question burst like soap bubbles, higher-percentage alcohols (e.g. ethanol, isopropanol ) can be used for disinfection: Bacteria and fungal cells are denatured Their membrane proteins and the perforation of their cell membrane are irreversibly inactivated, accordingly, enveloped viruses are also deprived of their lipid envelope , in which the docking proteins are located, at the same time as the proteins are denatured.
Denaturation by pure water
Proteins are in their natural environment in the presence of other proteins, dissolved salts , cofactors or metabolites that stabilize the natural protein structure in a more or less complex way. If salts and other smaller molecules are removed by dialysis of a protein solution against double-distilled water - preferably in the cold - one can often achieve selective (and reversible) denaturation, especially of large proteins which are precipitated under these conditions (precipitate).
Denaturation through modification and crosslinking
The use of molecular markings , fixation solutions and tanning agents , covalent crosslinkers (e.g. formaldehyde , paraformaldehyde or glutaraldehyde ) and solutions of heavy metal ions that form stable complexes occasionally change the catalytic center or a binding site of a protein in such a way that some functions are no longer fulfilled become. The protein is not unfolded (as is the case with thermal, chaotropic or pH-dependent denaturations), but it can be changed or fixed in a non-native conformation and lose functions. If the necessary functions of the protein remain unaffected by the fixation, other properties such as the biological half-life can also be changed by cross-linking . In the course of antigen unmasking , an attempt is made to reverse the effects of the fixation.
Denaturation in living things
Proteins are partially unfolded in cells before membrane transport by chaperones and then fold back. The base pairing of DNA is broken in sections by various DNA-binding proteins , e.g. B. in replication or transcription . The location of the start of denaturation is known as the denaturation bubble and is described in the Poland-Scheraga model . However, the DNA sequence , stiffness and torsion are not included. The lifespan of a denaturation bubble is between one microsecond and one millisecond.
Renaturation
After a protein (e.g. an enzyme ) has been denatured during the purification from a protein mixture, a return of the protein to its native form is necessary in order to measure the biological activity. However, this only works with proteins whose native conformation also represents the energetically most favorable state under isotonic conditions, but not with metastable proteins. Renaturation after chemical denaturation can be achieved by slowly diluting the denaturant, accompanied by restoration of the cofactors and the isotonic environment. Other additives are sometimes used. A reconstitution can be done afterwards. In the event of thermal denaturation of nucleic acids, these hybridize again when the temperature drops. The rate of renaturation of the base pairing of nucleic acids increases with the proportion of correct base pairing.
Differentiation from other changes
The structural changes mediated by proteins are not referred to as denaturation:
- molecules synthesized , transformed or degraded by enzymes , activation and deactivation reactions; for example, milk protein is coagulated by rennet enzymes
- Conformational changes due to chaperones or prions .
At very high temperatures, covalent bonds can also split and thus chain breaks ( depolymerization ). Such changes in the primary structure are not counted as denaturations. Likewise, acids such as alkalis can lead to the cleavage of covalent bonds at high concentrations and reaction temperatures. The primary structure then changes through hydrolysis . Such changes in the primary structure are normal chemical reactions and are not included in denaturation.
A borderline case is the cleavage of disulfide bridges between two protein strands. Although a covalent chemical bond is broken, the amino acid sequence in each individual strand is retained, which is why such a reductive cleavage of disulfide bridges, which in principle is reversible, is a denaturation.
Applications
In animals, proteins are denatured in the stomach .
Denaturation is used, among other things, in the production of protein-containing foods , e.g. B. when cooking or making panir or tofu . The denaturation during cooking makes digestion easier, in addition to killing microorganisms in the food and inactivating viruses . Various methods of disinfection and sometimes also of food preservation (when heating or drying) use the denaturation of proteins from undesired microorganisms, which kills them. In creating a permanent wave , hair is denatured to change its shape.
In biochemistry, denaturation is used to unfold proteins, e.g. B. chemically in SDS-PAGE or protein sequencing . When molecules bind to a protein, the thermal stability of the protein can be increased. The change in the denaturation temperature of a protein when another molecule binds can be measured with a thermal shift assay and the binding can thus be detected. For nucleic acids, PCR, DNA sequencing , Southern and Northern blot , in-situ hybridization , TGGE with thermal denaturation are used, while DGGE and urea - or formamide - polyacrylamide gel electrophoresis are carried out with chemical denaturation. Denaturation is also used to inactivate enzymes in a DNA extraction or an RNA extraction .
literature
- Friedrich Lottspeich , Haralabos Zorbas: Bioanalytics . Spektrum Akademischer Verlag, Heidelberg 1998, ISBN 978-3827400413 .
- Hubert Rehm , Thomas Letzel: The Experimenter: Protein Biochemistry / Proteomics . 6th edition, Spektrum Akademischer Verlag, Heidelberg 2009, ISBN 978-3827423122 .
Web links
- Animation: protein denaturation
- DFG Senate Commission SKLM: Safety assessment of the high pressure method (PDF; 188 kB)
Individual evidence
- ↑ Jane H. Dyson, Peter E. Wright: Intrinsically unstructured proteins and their functions In: Nature Reviews Molecular Cell Biology. (2005), Volume 6, Issue 3, pp. 197-208. doi: 10.1038 / nrm1589 .
- ↑ Hans-Dieter Jakubke, Hans Jeschkeit: Amino acids, peptides, proteins , Verlag Chemie, Weinheim, 402, 1982, ISBN 3-527-25892-2 .
- ↑ a b c d e Philipp Christen, Rolf Jaussi, Roger Benoit: Biochemistry and Molecular Biology. Springer-Verlag, 2015, ISBN 978-3-662-46430-4 , pp. 36, 37.
- ↑ CB Anfinsen : Principles that govern the folding of protein chains. In: Science. Volume 181, Number 4096, July 1973, pp. 223-230, PMID 4124164 .
- ↑ Yoshinori Mine, Tatsushi Noutomi, Noriyuki Haga: Thermally induced changes in egg white protein. In: Journal of Agricultural and Food Chemistry. 38, 1990, p. 2122, doi : 10.1021 / jf00102a004 .
- ↑ Charles Tanford : Protein denaturation. In: Advances in protein chemistry. Volume 23, 1968, pp. 121-282, PMID 4882248 .
- ↑ a b H. Robert Horton et al .: Biochemistry. Pearson Deutschland GmbH, 2008, ISBN 978-3-827-37312-0 , pp. 144-146.
- ↑ Hans-Dieter Jakubke, Hans Jeschkeit: amino acids, peptides, proteins , Verlag Chemie, Weinheim, 403, 1982, ISBN 3-527-25892-2 .
- ^ Yuan et al .: Shear-Stress-Mediated Refolding of Proteins from Aggregates and Inclusion Bodies. In: ChemBioChem . No. 16, 2015, pp. 393-396.
- ^ A b Peter Karlson: Karlsons Biochemie und Pathobiochemie. Georg Thieme Verlag, 2005, ISBN 978-3-133-57815-8 , p. 35.
- ^ Gerhard Richter: Practical Biochemistry. Georg Thieme Verlag, 2003, ISBN 978-3-131-32381-1 , pp. 144, 195.
- ↑ E. Palou, A. Lopet-Malo, GV Barbosa-Canovas and BG Swanson: High Pressure Treatment in Food Preservation , Marcel Dekker, New York., 1999
- ↑ Hans-Dieter Jakubke, Hans Jeschkeit: amino acids, peptides, proteins , Verlag Chemie, Weinheim, 403-404, 1982, ISBN 3-527-25892-2 .
- ↑ JP López-Alonso, M. Bruix, J. Font, M. Ribó, M. Vilanova, MA Jiménez, J. Santoro, C. González, DV Laurents: NMR spectroscopy reveals that RNase A is chiefly denatured in 40% acidic acid : implications for oligomer formation by 3D domain swapping. In: Journal of the American Chemical Society. Volume 132, Number 5, February 2010, pp. 1621-1630, doi : 10.1021 / ja9081638 , PMID 20085318 .
- ↑ Popova, E .: Phase equilibria in the precipitation of proteins from aqueous solutions. 2007, accessed November 20, 2019 .
- ↑ J. Marmur, PO Ts'O: Denaturation of deoxyribonucleic acid by formamide. In: Biochimica et Biophysica Acta . Volume 51, July 1961, pp. 32-36, PMID 13767022 .
- ↑ a b c d Academic Press: PROG NUCLEIC ACID RES & MOLECULAR BIO. Academic Press, 1963, ISBN 978-0-080-86289-7 , p. 267.
- ↑ a b Hyone-Myong Eun: Enzymology Primer for Recombinant DNA Technology. Elsevier, 1996, ISBN 978-0-080-53113-7 , p. 67.
- ↑ a b X. Wang, HJ Lim, A. Son: Characterization of denaturation and renaturation of DNA for DNA hybridization. In: Environmental health and toxicology. Volume 29, 2014, p. E2014007, doi : 10.5620 / eht.2014.29.e2014007 , PMID 25234413 , PMC 4168728 (free full text).
- ↑ François Sicard, Nicolas Destainville, Manoel Manghi: DNA denaturation bubbles: Free-energy landscape and nucleation / closure rates . In: The Journal of Chemical Physics . 142, No. 3, January 21, 2015, p. 034903. arxiv : 1405.3867 . doi : 10.1063 / 1.4905668 .
- ^ Simon Lieu: The Poland-Scheraga Model. (2015): 0-5. Massachusetts Institute of Technology, May 14, 2015.
- ^ C. Richard, AJ Guttmann: Poland – Scheraga Models and the DNA Denaturation Transition. In: Journal of Statistical Physics. 115, 2004, p. 925, doi : 10.1023 / B: JOSS.0000022370.48118.8b .
- ↑ Grégoire Altan-Bonnet, Albert Libchaber, Oleg Krichevsky: Bubble Dynamics in Double-Stranded DNA . In: Physical Review Letters . 90, No. 13, April 1, 2003. doi : 10.1103 / physrevlett.90.138101 .
- ↑ Winfried Storhas: Bioprocess development. John Wiley & Sons, 2013, ISBN 978-3-527-67385-8 , p. 621.
- ^ Rolf Knippers: Molecular Genetics. Georg Thieme Verlag, 2015, ISBN 978-3-131-68330-4 .
- ^ Brockhaus ABC Chemie , VEB FA Brockhaus Verlag Leipzig 1965, p. 274.
- ^ Robert Ebermann: Textbook food chemistry and nutrition. Springer-Verlag, 2011, ISBN 978-3-709-10211-4 , p. 57.
- ^ Peter W. Atkins: Physical chemistry. John Wiley & Sons, 2006, ISBN 978-3-527-31546-8 , p. 754.