Molecular marking
The molecular marking ( English labeling , labeling , tagging ) is a method of chemistry and biochemistry for the selective binding of an atom or molecule to a molecule. The marking is used for further tracking of the marked molecule or to elucidate a reaction mechanism ( English crossover experiment ).
properties

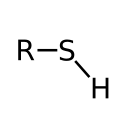
Depending on their characteristic functional groups, molecules can be provided with a selectively binding label that carries a traceable signal molecule ( reporter molecule ). The label consists of a binding group (coupling group), occasionally a connector (linker) and a detectable molecule (reporter). The marking is mostly covalently linked via the coupling group. In the case of nuclides , however, it can also be bound ionically and, in the case of indirect detection, via a mixture of ionic bonds, van der Waals bonds , hydrophobic effects , hydrogen bonds and sometimes also disulphide bonds . In contrast to staining with most selectively binding protein dyes or nucleic acid dyes, biomolecules are covalently labeled either on individual atoms or in a sequence-specific manner.
Reporter types
The reporter molecules are generally about selectively reactive coupling groups covalently linked to a flexible spacer ( English left , connector ') coupled to certain functional groups of the molecule to be labeled. Nuclides are a special case, because of their comparatively small molecule size, they can also be coupled directly to another molecule (without a reactive coupling group or a linker). While isotope labeling is mostly used to label small molecules , many different types of reporter have been developed for labeling various biopolymers such as proteins and DNA.
As reporter molecules in marks are nuclides ( radioactive and non-radioactive), biotin , reporter enzymes , oligonucleotides , fluorophores (as part of a fluorescent label ) or proteins and protein tags used.
Coupling types
The markings with nuclides have the smallest changes in molar mass among the markings and therefore lead to a comparatively smaller change in the protein structure and biological activity , but the radioactive nuclides are accompanied by increased safety measures and requirements for the safety laboratory. Nuclides ( isotopes and isobars ) can be directly covalently coupled to the molecule to be labeled ( chemical coupling ), besides being covalently attached via a coupling group, ionically coupled (e.g. via chelators such as DTPA , TTHA or chelating peptides such as protein -Tag His-Tag ) by a chemical total synthesis (e.g. by peptide synthesis, by phosphoramidite synthesis) or in a biosynthesis by the conversion of nuclide-labeled precursor substances ( metabolic labeling ). In the course of chemical isotope labeling or metabolic isotope labeling, molecules with radioactive isotopes (e.g. 3 H, 11 C, 13 C, 14 C, 13 N, 15 O, 18 F, 26 Al, 32 P, 33 P, 35 S, 36 Cl, 41 Ca, 125 I, 131 I). The isotopes of elements that do not naturally occur in biomolecules such as iodine , 99 technetium or 113 indium must first be chemically coupled to the precursor molecules in the case of metabolic labeling.
The other types of reporter molecules can be covalently attached via a coupling group, generated by a chemical total synthesis or in a biosynthesis. The biosynthesis takes place in the metabolism through the conversion of reporter-marked precursor substances ( metabolic marking ). With high selectivity and low toxicity , labeling can also take place partly metabolically and partly synthetically, as in the case of bioorthogonal labeling . Metabolically incorporated precursor substances are then used after protein purification or DNA purification for the subsequent chemical coupling of a reporter molecule using the selectively reactive group, e.g. B. via click chemistry (1,3-dipolar cycloaddition with azides and cyclooctynes , a copper-free click chemistry), via cycloaddition between nitrones and cyclooctynes, via oxime / hydrazone formation from aldehydes or ketones , via tetrazine ligation, by isonitrile-based click reaction, by quadricyclane ligation and by Staudinger reaction between azides and triarylphosphines.
For tracking the time course of a labeled molecule in the context of metabolism is a variant of metabolic labeling, the pulse-labeling ( English pulse labeling ) was used. In the case of pulse marking, the marking period is limited in time, so that the marking of the molecule and its metabolites can be more precisely delimited and the change in marking to individual subsequent substances in a metabolic pathway can be observed. The tracking multiple metabolites is also known as metabolic flux analysis (at the same English metabolic flux analysis indicated). Through the molecular marking , the cell membranes of whole cells can also be marked by marking surface proteins and / or the glycocalyx , e.g. B. for flow cytometry , fluorescence microscopy or fluorescence tomography .
In a protection assay , markers are used, among other things, to mark uncovered areas on the surface of a molecule, whereby the surface or the binding site of a binding molecule can be determined. The areas of a protein accessible for marking are an indication that the marked amino acids are freely accessible on the protein surface. Folded areas of proteins and areas that have bound another molecule are less amenable to labeling.
Direct and indirect evidence
In the case of a molecular marking, the molecule to be detected can either be provided directly with a reporter molecule or indirectly detected by selectively binding reporter-carrying molecules. Molecular labeling methods are:
- Direct molecular markings
- chemical total synthesis (only in vitro )
- chemical coupling ( in vitro coupling of the reporter molecule)
- bioorthogonal marking ( in vitro coupling of the reporter molecule)
- metabolic labeling in vitro (e.g. PCR, in vitro translation)
- metabolic labeling in vivo (feeding labeled molecules)
- Reporter proteins
- Protein tags or RNA tags that bind a reporter molecule.
- Indirect molecular markings
- Immune labeling (for proteins, protein tags and carbohydrates)
- Lectin marking (for carbohydrates and glycoproteins)
- Hybridization probes (for nucleic acids)
- Biotinylation
- Reporter genes
Protein labeling
Reactive groups in proteins
In contrast to carbohydrates and nucleic acids, some of the structural motifs of the biomolecules only occur in proteins due to the different amino acids they contain , e.g. B. sulfhydryl -containing cysteines or phenol residues in tyrosines . These structural motifs can be selectively marked with appropriate coupling reagents. Strategies are being investigated to mark only one of several amino acids of the same type.
coupling
Cysteines can with maleimides , disulfides , iodoacetamide (eg. IAEDANS ), Haloacetylen , aziridines , Acryloylen , arylating agents , vinyl sulphones , pyridyl - and other disulfides react selectively. Amino acids with primary amines on the side chain such as lysine can be labeled by succinimide esters ( N -hydroxysuccinimide , sulfosuccinimide or other succinimidyl esters) or certain isothiocyanates such as PITC or FITC . Carboxy groups can be coupled to amine groups by activation with carbodiimides . After oxidation of proteins or by reductive alkylation , various reporter molecules can be coupled. Nucleophilic groups in proteins can be coupled via tosylation . With enzymes, proteins can often be labeled with an increased selectivity on protein tags, e.g. B. by Sortase , by Transglutaminase , by Haloalkandehalogenase or by Phosphopantetheinyltransferase. The crosslinking and fixation of various amino acid side chains in proteins is based on the same principle as labeling by coupling .
Photoreactive molecules (e.g. aryl azides , diazirines ) can also be used as a reactive group of a label on proteins in order to be able to better control the time of coupling, since the reaction is only triggered with UV radiation, e.g. B. in the course of photoaffinity labeling . Because of the lower selectivity of these free-radical coupling groups, the functionality of the protein is often reduced by reaction of the free-radical coupling group with important functions (e.g. an active center of an enzyme or a binding site ). An advantage of radical couplings, on the other hand, is the independence from the occurrence of certain amino acids in the protein to be coupled. Therefore, photoreactive markings are mostly used when no or only one amine or sulfhydryl group is available for selective coupling or when subsequent functionality is insignificant. There are also photoreactive diazirine or azide-containing analogs of the amino acids leucine ( photo-leucine ), methionine and p-benzoyl- phenylalanine , which can be incorporated into the protein during translation. By peptide synthesis or in vitro translation can peptides with labeled amino acid derivatives are prepared.
Some protease inhibitors lead to selective labeling of proteins. In a label transfer experiment, a cleavable cross-link with a reporter between two neighboring molecules is used in order to transfer the reporter molecule between these molecules by cleaving the cross-link and thereby to prove their proximity. The cleavable crosslinker used here contains the reporter group between the cleavage point of the crosslinker and the coupling group for the mostly unknown binding target molecule. In the DIGE , differently labeled samples are separated together using SDS-PAGE or 2D gel electrophoresis . In a proximity ligation assay , the neighborhood of oligonucleotide-labeled proteins is detected by PCR. Proteins can be labeled with oligonucleotides for detection by hybridization. Proteins can also be biotinylated in vitro with biotin succinimidyl esters or provided with a protein tag with biotinylation (e.g. Avi-Tag, BCCP-Tag, Strep-Tag) in vivo and then indirectly labeled with avidin or streptavidin conjugates become.
Nuclide markings
Nuclide labeling mostly uses radioactive nuclides due to the comparatively high sensitivity (low detection limit ) and the simplicity of detection by autoradiography , by scintillation counter or by positron emission tomography . In nuclear magnetic resonance spectroscopy and mass spectrometry , non-radioactive nuclides can also be used.
The nuclide labeling is done either chemically or biosynthetically. The biosynthetic labeling takes place z. B. as metabolic labeling by feeding cell cultures or test animals with labeled precursor molecules. By radioiodination, tyrosines can be labeled in vitro with radioactive iodine . Phosphorylations of serine , threonine and tyrosine are usually carried out enzymatically with suitable protein kinases and radioactive phosphorus-containing adenosine triphosphate . A radioactively labeled prenylation can be carried out with 3 H- mevalonic acid . In addition, the C-terminus is methylated in the course of a prenylation , which can be reversibly radiolabeled by 3 H-labeled S-adenosyl-methionine . N-terminal labeling can be carried out by myristylation . A geranylgeranylation can be performed with Azidogeranylen. Sulphation can be detected with 35 S-labeled sulphate. Hydrogen atoms can by a deuterium with deuterium to be replaced. Marking with radioactive isotopes allows tracking by autoradiography . In mass spectrometry, isobaric labeling is used to differentiate between different samples. In addition to the normally used nuclides 1 hydrogen or 15 nitrogen , markings with 13 carbon or 19 fluorine are used in NMR spectroscopy .
Bioorthogonal markings
Proteins can be labeled bioorthogonally . Various selective coupling reactions are used here, e.g. B. via Sonogashira coupling , via copper-catalyzed alkyne-azide cycloaddition , via Heck reaction , via oxime ligation, via cyclopropene-azide coupling, via myristylation, via cyanobenzothiazole condensation or via tetrazole-alkene cycloaddition. After biosynthesis, membrane proteins can be enzymatically coupled with azides , which in turn are coupled with reporter molecules via the Staudinger reaction.
Recombinant Tags
Proteins can be produced as recombinant proteins with a protein tag or with a reporter protein as fusion protein, which are attached N-terminally or C-terminally to the protein during translation . A protein can be purified or detected via the protein tag inserted into the recombinant protein , e.g. B. with GFP or with reporter enzymes . Some protein tags are subsequently coupled bioorthogonally with a reporter molecule after the biosynthesis, e.g. B. the Snap-Tag , the Polyhistidine-Tag , the Flash-Tag , the ReAsH-Tag , the Flag-Tag , the Clip-Tag , the HyRe -Tag , the Beta-Lactamase-Tag , the LAP-Tag and the Sortase day. Proteins can be marked post-translationally by inteins .
Indirect markings
In the case of an immunolabeling with immunoconjugates , the selectivity of the marking is based on the binding of antibodies . In addition, the antibody is usually marked with a reporter molecule itself or via a secondary antibody, which is used indirectly for detection, e.g. B. in fluorescence microscopy, Western blot and ELISA . The epitope bound by the antibody is either in the protein or on a protein tag. Proteins can also be detected indirectly using reporter genes.
Nucleic acid labeling
Reactive groups in nucleic acids
Compared to proteins, DNA has a lower number of available functional groups due to the nucleotides used , including an amino group in the nucleobase adenine that can be used for coupling .
Coupling types
The phosphate group can be marked by radioactive phosphate with a 32 phosphorus atom in vitro in the course of random priming , nick translation , generation by phosphoramidite synthesis or in vivo by biosynthesis with radioactive phosphate. DNA can also be labeled bioorthogonally, e.g. B. with 5-ethynyl-2'dUTP. The amino group of adenine can react in vitro with succinimidyl esters or isothiocyanates.
A polymerase chain reaction can label DNA in vitro with unnatural nucleotide analogs , e.g. B. BrdU , digoxigenin -dUTP, hydroxymethyl-dCTP and fluorescent nucleotides in DNA sequencing according to Sanger, QPCR or in-situ hybridization with hybridization probes .
RNA
RNA can be labeled biosynthetically with RNA tags, chemically or with hybridization probes, among other things. In the case of RNA tags, DNA sequences of aptamers are usually cloned into the open reading frame of a gene . After the RNA has been generated with the RNA tag by transcription , secondary structures are formed . B. bind selectively to dextran , streptavidin or dyes .
Carbohydrate marking

Marked carbohydrates are generated by metabolic or bioorthogonal marking, chemically marked or the carbohydrates are detected indirectly via lectins .
literature
- Hubert Rehm : Protein Biochemistry / Proteomics, The Experimentator . 5th edition. Spektrum Akademischer Verlag, Heidelberg 2006, ISBN 3-8274-1726-0 .
- Friedrich Lottspeich , Haralabos Zorbas: Bioanalytics . Spektrum Akademischer Verlag, Heidelberg 1998, ISBN 3-8274-0041-4 .
- Juan S. Bonifacino: Protein Labeling and Immunoprecipitation. In: Current Protocols in Cell Biology . Wiley-VCH 1998. doi: 10.1002 / 0471143030.cb0700s15 . PDF .
Individual evidence
- ↑ MJ Hinner, K. Johnsson: How to obtain labeled proteins and what to do with them. In: Current Opinion in Biotechnology. Volume 21, number 6, December 2010, pp. 766-776, doi: 10.1016 / j.copbio.2010.09.011 . PMID 21030243 .
- ^ R. Wombacher, VW Cornish: Chemical tags: applications in live cell fluorescence imaging. In: Journal of biophotonics. Volume 4, number 6, June 2011, pp. 391-402, doi: 10.1002 / jbio.201100018 . PMID 21567974 .
- ^ HM O'Hare, K. Johnsson, A. Gautier: Chemical probes shed light on protein function. In: Current opinion in structural biology. Volume 17, Number 4, August 2007, pp. 488-494, doi: 10.1016 / j.sbi.2007.07.005 . PMID 17851069 .
- ^ LW Miller, VW Cornish: Selective chemical labeling of proteins in living cells. In: Current opinion in chemical biology. Volume 9, Number 1, February 2005, pp. 56-61, doi: 10.1016 / j.cbpa.2004.12.007 . PMID 15701454 .
- ↑ DR Reddy, LE Pedró Rosa, LW Miller: Luminescent trimethoprim-polyaminocarboxylate lanthanide complex conjugates for selective protein labeling and time-resolved bioassays. In: Bioconjugate Chemistry . Volume 22, Number 7, July 2011, pp. 1402-1409, doi: 10.1021 / bc200131k . PMID 21619068 . PMC 3140616 (free full text).
- ↑ N. Maindron, S. Poupart, M. Hamon, JB Langlois, N. Plé, L. Jean, A. Romieu, PY Renard: Synthesis and luminescence properties of new red-shifted absorption lanthanide (III) chelates suitable for peptide and protein labeling. In: Organic & biomolecular chemistry. Volume 9, Number 7, April 2011, pp. 2357-2370, doi: 10.1039 / c0ob00832j . PMID 21321764 .
- ↑ G. Licini, P. Scrimin: Metal-ion-binding peptides: from catalysis to protein tagging. In: Angewandte Chemie. Volume 42, Number 38, October 2003, pp. 4572-4575, doi: 10.1002 / anie.200301668 . PMID 14533147 .
- ↑ a b G. H. Müller: Protein labeling with 3H-NSP (N-succinimidyl- [2,3-3H] propionate). In: Journal of cell science. Volume 43, June 1980, pp. 319-328, PMID 7419623 .
- ↑ B. Långström, F. Karimi, Y. Watanabe: Endogenous compounds labeled with radionuclides of short half-life-some perspectives. In: Journal of Labeled Compounds and Radiopharmaceuticals . Volume 56, number 3-4, 2013 Mar-Apr, pp. 251-262, doi: 10.1002 / jlcr.3033 . PMID 24285332 .
- ^ G. van Hall: Correction factors for 13C-labeled substrate oxidation at whole-body and muscle level. In: The Proceedings of the Nutrition Society. Volume 58, Number 4, November 1999, pp. 979-986, PMID 10817166 .
- ↑ a b c d M. Palmblad, BA Buchholz, DJ Hillegonds, JS Vogel: Neuroscience and accelerator mass spectrometry. In: Journal of Mass Spectrometry . Volume 40, Number 2, February 2005, pp. 154-159, doi: 10.1002 / jms.734 . PMID 15706618 .
- ↑ a b c P. W. Miller, NJ Long, R. Vilar, AD Gee: Synthesis of 11C, 18F, 15O, and 13N radiolabels for positron emission tomography. In: Angewandte Chemie. Volume 47, number 47, 2008, pp. 8998-9033, doi: 10.1002 / anie.200800222 . PMID 18988199 .
- ↑ a b A. M. Tolkovsky, A. Wyttenbach: Differential phosphoprotein labeling (DIPPL) using 32P and 33P. In: Methods in molecular biology. Volume 527, 2009, pp. 21-9, xi, doi : 10.1007 / 978-1-60327-834-8_2 . PMID 19241002 .
- ↑ MA Lydan, DH O'Day: Production of 35S-labeled proteins in E. coli and their use as molecular probes. In: Methods in molecular biology. Volume 31, 1994, pp. 389-396, doi: 10.1385 / 0-89603-258-2: 389 . PMID 7921035 .
- ↑ FW Fitch, J. WINEBRIGHT, PV HARPER: Iodine-125 as a protein label in immunology. In: Science. Volume 135, Number 3508, March 1962, pp. 1068-1069, PMID 13893326 .
- ↑ LR MELCHER, SP MASOUREDIS: The in vivo stability of the I131 protein label of rabbit antibody in guinea pigs as determined by the quantitative precipitin reaction. In: Journal of Immunology (Baltimore, Md.: 1950). Volume 67, Number 5, November 1951, pp. 393-402, PMID 14898075 .
- ↑ C. Xavier, N. Devoogdt, S. Hernot, I. Vaneycken, M. D'Huyvetter, J. De Vos, S. Massa, T. Lahoutte, V. Caveliers: Site-specific labeling of his-tagged Nanobodies with 99mTc: a practical guide. In: Methods in molecular biology. Volume 911, 2012, pp. 485-490, doi : 10.1007 / 978-1-61779-968-6_30 . PMID 22886271 .
- ↑ MH Adatepe, P. Penkoske, A. Van Amberg, T. Wharton, RG Evens, EJ Potchen: Red cell and plasma protein labeling with 113 m In. In: The International journal of applied radiation and isotopes. Volume 22, Number 8, August 1971, pp. 498-501, PMID 5097086 .
- ^ JA Prescher, DH Dube, Carolyn Bertozzi : Chemical remodeling of cell surfaces in living animals. In: Nature . Volume 430, number 7002, August 2004, pp. 873-877, doi: 10.1038 / nature02791 . PMID 15318217 .
- ^ JA Prescher, CR Bertozzi: Chemistry in living systems. In: Nature Chemical Biology . Volume 1, Number 1, June 2005, pp. 13-21, doi: 10.1038 / nchembio0605-13 . PMID 16407987 .
- ^ AB Neef, NW Luedtke: Dynamic metabolic labeling of DNA in vivo with arabinosyl nucleosides. In: Proceedings of the National Academy of Sciences . Volume 108, number 51, December 2011, pp. 20404-20409, doi: 10.1073 / pnas.1101126108 . PMID 22143759 . PMC 3251089 (free full text).
- ↑ HC Kolb, MG Finn, KB Sharpless: Click Chemistry: Diverse Chemical Function from a Few Good Reactions. In: Angewandte Chemie. Volume 40, Number 11, June 2001, pp. 2004-2021, PMID 11433435 .
- ↑ a b A. Cieslar-Pobuda, MJ Los: Prospects and limitations of “Click Chemistry” -based DNA labeling technique employing 5-ethynyl-2'deoxyuridine (EdU). In: Cytometry. Part A: the journal of the International Society for Analytical Cytology. Volume 83, number 11, November 2013, pp. 977-978, doi: 10.1002 / cyto.a.22394 . PMID 24115755 .
- ↑ JM Baskin, JA Prescher, ST Laughlin, NJ Agard, PV Chang, IA Miller, A. Lo, JA Codelli, CR Bertozzi: Copper-free click chemistry for dynamic in vivo imaging. In: Proceedings of the National Academy of Sciences . Volume 104, Number 43, October 2007, pp. 16793-16797, doi: 10.1073 / pnas.0707090104 . PMID 17942682 , PMC 2040404 (free full text).
- ↑ Xinghai Ning, Rinske P. Temming and a .: Protein Modification by Strain-Promoted Alkyne-Nitrone Cycloaddition. In: Angewandte Chemie International Edition. 49, 2010, pp. 3065-3068, doi: 10.1002 / anie.201000408 .
- ↑ KJ Yarema, LK Mahal, RE Bruehl, EC Rodriguez, CR Bertozzi: Metabolic Delivery of Ketone Groups to Sialic Acid Residues. APPLICATION TO CELL SURFACE GLYCOFORM ENGINEERING . In: Journal of Biological Chemistry . 273, No. 47, 1998, pp. 31168-79. doi : 10.1074 / jbc.273.47.31168 . PMID 9813021 .
- ↑ Melissa L. Blackman, Maksim Royzen, Joseph M. Fox: The Tetrazine Ligation: Fast Bioconjugation based on Inverse-electron-Demand Diels-Alder Reactivity . In: Journal of the American Chemical Society . 130, No. 41, 2008, pp. 13518-9. doi : 10.1021 / ja8053805 . PMID 18798613 . PMC 2653060 (free full text).
- ^ Henning Stöckmann, André A. Neves, Shaun Stairs, Kevin M. Brindle, Finian J. Leeper: Exploring isonitrile-based click chemistry for ligation with biomolecules . In: Organic & Biomolecular Chemistry . 9, No. 21, 2011, p. 7303. doi : 10.1039 / C1OB06424J .
- ↑ Ellen M. Sletten, Carolyn R. Bertozzi: A Bioorthogonal Quadricyclane Ligation . In: Journal of the American Chemical Society . 133, No. 44, 2011, pp. 17570-3. doi : 10.1021 / ja2072934 . PMID 21962173 . PMC 3206493 (free full text).
- ^ E. Saxon, CR Bertozzi: Cell surface engineering by a modified Staudinger reaction. In: Science. Volume 287, Number 5460, March 2000, pp. 2007-2010, PMID 10720325 .
- ^ NJ Agard, JM Baskin, JA Prescher, A. Lo, CR Bertozzi: A comparative study of bioorthogonal reactions with azides. In: ACS chemical biology. Volume 1, Number 10, November 2006, pp. 644-648, PMID 17175580 .
- ^ SH Weisbrod, A. Baccaro, A. Marx: Site-specific DNA labeling by Staudinger ligation. In: Methods in molecular biology. Volume 751, 2011, pp. 195-207, doi : 10.1007 / 978-1-61779-151-2_12 . PMID 21674332 .
- ^ J. O'Grady, J. Schwender, Y. Shachar-Hill, JA Morgan: Metabolic cartography: experimental quantification of metabolic fluxes from isotopic labeling studies. In: Journal of experimental botany. Volume 63, Number 6, March 2012, pp. 2293-2308, doi: 10.1093 / jxb / ers032 . PMID 22371075 . PDF .
- ^ MA Orman, F. Berthiaume, IP Androulakis, MG Ierapetritou: Advanced stoichiometric analysis of metabolic networks of mammalian systems. In: Critical reviews in biomedical engineering. Volume 39, Number 6, 2011, pp. 511-534, PMID 22196224 . PMC 3634616 (free full text).
- ↑ SB Crown, MR Antoniewicz: Parallel labeling experiments and metabolic flux analysis: Past, present and future methodologies. In: Metabolic engineering. Volume 16, March 2013, pp. 21-32, doi: 10.1016 / j.ymben.2012.11.010 . PMID 23246523 .
- ↑ X. Chen, Y. Shachar-Hill: Insights into metabolic efficiency from flux analysis. In: Journal of experimental botany. Volume 63, Number 6, March 2012, pp. 2343-2351, doi: 10.1093 / jxb / ers057 . PMID 22378949 . PDF .
- ^ WH So, Y. Zhang, W. Kang, CT Wong, H. Sun, J. Xia: Site-selective covalent reactions on proteinogenic amino acids. In: Current Opinion in Biotechnology. Volume 48, December 2017, pp. 220-227, doi : 10.1016 / j.copbio.2017.06.003 , PMID 28688251 .
- ↑ IM Riederer, RM Herrero, G. Leuba, BM Riederer: Serial protein labeling with infrared maleimide dyes to identify cysteine modifications. In: Journal of proteomics. Volume 71, number 2, July 2008, pp. 222-230, doi: 10.1016 / j.jprot.2008.04.006 . PMID 18556256 .
- Jump up ↑ J. Guy, R. Castonguay, NB Campos-Reales Pineda, V. Jacquier, K. Caron, SW Michnick, JW Keillor: De novo helical peptides as target sequences for a specific, fluorogenic protein labeling strategy. In: Molecular bioSystems. Volume 6, Number 6, June 2010, pp. 976-987, doi: 10.1039 / b918205e . PMID 20485742 .
- ^ H. Kratz, A. Haeckel, R. Michel, L. Schönzart, U. Hanisch, B. Hamm, E. Schellenberger: Straightforward thiol-mediated protein labeling with DTPA: Synthesis of a highly active 111In-annexin A5-DTPA tracer . In: EJNMMI research. Volume 2, number 1, 2012, p. 17, doi: 10.1186 / 2191-219X-2-17 . PMID 22541756 . PMC 3444359 (free full text).
- ↑ KK Han, A. Delacourte, B. Hemon: Chemical modification of thiol group (s) in protein: application to the study of anti-microtubular drugs binding. In: Comparative biochemistry and physiology. B, Comparative biochemistry. Volume 88, Number 4, 1987, pp. 1057-1065, PMID 3322663 .
- ^ RA Evangelista, A. Pollak, B. Allore, EF Templeton, RC Morton, EP Diamandis: A new europium chelate for protein labeling and time-resolved fluorometric applications. In: Clinical biochemistry. Volume 21, Number 3, June 1988, pp. 173-178, PMID 3390907 .
- ↑ J. Roeser, R. Bischoff, AP Bruins, HP Permentier: Oxidative protein labeling in mass-spectrometry-based proteomics. In: Analytical and bioanalytical chemistry. Volume 397, Number 8, August 2010, pp. 3441-3455, doi: 10.1007 / s00216-010-3471-8 . PMID 20155254 . PMC 2911539 (free full text).
- ↑ N. Jentoft, DG Dearborn: Protein labeling by reductive alkylation. In: Methods in enzymology. Volume 91, 1983, pp. 570-579, PMID 6855602 .
- ↑ S. Tsukiji, M. Miyagawa, Y. Takaoka, T. Tamura, I. Hamachi: Ligand-directed tosyl chemistry for protein labeling in vivo. In: Nature chemical biology. Volume 5, Number 5, May 2009, pp. 341-343, doi: 10.1038 / nchembio.157 . PMID 19330012 .
- ^ A b T. Matsumoto, R. Takase, T. Tanaka, H. Fukuda, A. Kondo: Site-specific protein labeling with amine-containing molecules using Lactobacillus plantarum sortase. In: Biotechnology journal. Volume 7, number 5, May 2012, pp. 642-648, doi: 10.1002 / biot.201100213 . PMID 21922670 .
- ↑ N. Kamiya, H. Abe, M. Goto, Y. Tsuji, H. Jikuya: Fluorescent substrates for covalent protein labeling catalyzed by microbial transglutaminase. In: Organic & biomolecular chemistry. Volume 7, Number 17, September 2009, pp. 3407-3412, doi: 10.1039 / b904046c . PMID 19675894 .
- ↑ GV lot LP encell, MG McDougall, DD Hartzell, N. Karassina, C. Zimprich, MG Wood, R. Learish, RF Ohana, M. Urh, D. Simpson, J. Mendez, K. Zimmerman, P. Otto , G. Vidugiris, J. Zhu, A. Darzins, DH Klaubert, RF Bulleit, KV Wood: HaloTag: a novel protein labeling technology for cell imaging and protein analysis. In: ACS chemical biology. Volume 3, Number 6, June 2008, pp. 373-382, doi: 10.1021 / cb800025k . PMID 18533659 .
- ↑ Y. Zou, J. Yin: Phosphopantetheinyl transferase catalyzed site-specific protein labeling with ADP conjugated chemical probes. In: Journal of the American Chemical Society. Volume 131, Number 22, June 2009, pp. 7548-7549, doi: 10.1021 / ja902464v . PMID 19441828 .
- ↑ J. Roeser, R. Bischoff, AP Bruins, HP Permentier: Oxidative protein labeling in mass-spectrometry-based proteomics. In: Analytical and bioanalytical chemistry. Volume 397, Number 8, August 2010, pp. 3441-3455, doi: 10.1007 / s00216-010-3471-8 . PMID 20155254 . PMC 2911539 (free full text).
- ^ M. Suchanek, A. Radzikowska, C. Thiele: Photo-leucine and photo-methionine allow identification of protein-protein interactions in living cells . In: Nature Methods . 2, No. 4, 2005, pp. 261-268. doi : 10.1038 / nmeth752 . PMID 15782218 .
- ↑ JW Chin, SW Santoro, AB Martin, DS King, L. Wang, PG Schultz: Addition of p-azido-L-phenylalanine to the genetic code of Escherichia coli. In: Journal of the American Chemical Society. Volume 124, Number 31, August 2002, pp. 9026-9027, PMID 12148987 .
- ↑ DF Winkler, PL McGeer: Protein labeling and biotinylation of peptides during spot synthesis using biotin p-nitrophenyl ester (biotin-ONp). In: Proteomics. Volume 8, number 5, March 2008, pp. 961-967, doi: 10.1002 / pmic.200700909 . PMID 18324722 .
- ↑ S. Mamaev, J. Olejnik, EK Olejnik, KJ Rothschild: Cell-free N-terminal protein labeling using initiator suppressor tRNA. In: Analytical biochemistry. Volume 326, number 1, March 2004, pp. 25-32, doi: 10.1016 / j . from 2003.11.002 . PMID 14769332 .
- ^ DA Fancy: Elucidation of protein-protein interactions using chemical cross-linking or label transfer techniques. In: Current opinion in chemical biology. Volume 4, Number 1, February 2000, pp. 28-33, PMID 10679368 .
- ↑ SS Andrews, ZB Hill, BG Perera, DJ Maly: Label transfer reagents to probe p38 MAPK binding partners. In: ChemBioChem . Volume 14, number 2, January 2013, pp. 209-216, doi: 10.1002 / cbic.201200673 . PMID 23319368 . PMC 3762675 (free full text).
- ↑ JB Denny, Günter Blobel : 125I-labeled crosslinking reagent that is hydrophilic, photoactivatable, and cleavable through an azo linkage. In: Proceedings of the National Academy of Sciences . Volume 81, Number 17, September 1984, pp. 5286-5290, PMID 6433347 . PMC 391688 (free full text).
- ↑ MM Shaw, BM Riederer: Sample preparation for two-dimensional gel electrophoresis. In: Proteomics. Volume 3, Number 8, August 2003, pp. 1408-1417, doi: 10.1002 / pmic.200300471 . PMID 12923765 .
- ↑ JF Timms, R. Cramer: Difference gel electrophoresis. In: Proteomics. Volume 8, number 23-24, December 2008, pp. 4886-4897, doi: 10.1002 / pmic.200800298 . PMID 19003860 .
- ↑ BM Riederer: Non-covalent and covalent protein labeling in two-dimensional gel electrophoresis. In: Journal of proteomics. Volume 71, number 2, July 2008, pp. 231–244, doi: 10.1016 / j.jprot.2008.05.001 . PMID 18556257 .
- ↑ O. Söderberg, M. Gullberg, M. Jarvius, K. Ridderstråle, KJ Leuchowius, J. Jarvius, K. Wester, P. Hydbring, F. Bahram, LG Larsson, U. Landegren: Direct observation of individual endogenous protein complexes in situ by proximity ligation. In: Nature methods. Volume 3, Number 12, December 2006, pp. 995-1000, doi: 10.1038 / nmeth947 . PMID 17072308 .
- ↑ DJ Hnatowich, G. Mardirossian, M. Rusckowski, P. Winnard: Protein labeling via deoxyribonucleic acid hybridization. In: Nuclear medicine communications. Volume 17, Number 1, January 1996, pp. 66-75, PMID 8692476 .
- ^ KH Lim, H. Huang, A. Pralle, S. Park: Stable, high-affinity streptavidin monomer for protein labeling and monovalent biotin detection. In: Biotechnology and bioengineering. Volume 110, number 1, January 2013, pp. 57-67, doi: 10.1002 / bit.24605 . PMID 22806584 .
- ↑ V. Tolmachev, A. Orlova: Influence of labeling methods on biodistribution and imaging properties of radiolabelled peptides for visualization of molecular therapeutic targets. In: Current medicinal chemistry. Volume 17, Number 24, 2010, pp. 2636-2655, PMID 20491631 .
- ↑ V. Tolmachev, S. Stone-Elander: Radiolabelled proteins for positron emission tomography: Pros and cons of labeling methods. In: Biochimica et Biophysica Acta . Volume 1800, number 5, May 2010, pp. 487-510, doi: 10.1016 / j.bbagen.2010.02.002 . PMID 20153401 .
- ↑ J. Meisenhelder, T. Hunter: Radioactive protein-labeling techniques. In: Nature. Volume 335, Number 6186, September 1988, p. 120, doi: 10.1038 / 335120a0 . PMID 3412469 .
- ↑ S. Caplan, M. Baniyash: Radioiodination of cellular proteins. In: Current protocols in cell biology / editorial board, Juan S. Bonifacino ... [et al.]. Chapter 7, August 2002, S. Unit 7.10, doi: 10.1002 / 0471143030.cb0710s15 . PMID 18228409 .
- ↑ M. Sunbul, J. Yin: Site specific protein labeling by enzymatic posttranslational modification. In: Organic & biomolecular chemistry. Volume 7, Number 17, September 2009, pp. 3361-3371, doi: 10.1039 / b908687k . PMID 19675886 .
- ↑ a b c s. Bonifacino
- ^ A b W. P. Heal, MH Wright, E. Thinon, EW Tate: Multifunctional protein labeling via enzymatic N-terminal tagging and elaboration by click chemistry. In: Nature protocols. Volume 7, number 1, January 2012, pp. 105–117, doi: 10.1038 / nprot.2011.425 . PMID 22193303 .
- ^ MW Rose, J. Xu, TA Kale, G. O'Doherty, G. Barany, MD Distefano: Enzymatic incorporation of orthogonally reactive prenylazide groups into peptides using geranylazide diphosphate via protein farnesyltransferase: implications for selective protein labeling. In: Biopolymers. Volume 80, number 2-3, 2005, pp. 164-171, doi: 10.1002 / bip.20239 . PMID 15810014 .
- ↑ KM Coombs: Quantitative proteomics of complex mixtures. In: Expert review of proteomics. Volume 8, number 5, October 2011, pp. 659-677, doi: 10.1586 / epr.11.55 . PMID 21999835 .
- ^ Y. Xu, S. Matthews: TROSY NMR spectroscopy of large soluble proteins. In: Topics in current chemistry. Volume 335, 2013, pp. 97-119, doi : 10.1007 / 128_2011_228 . PMID 21928013 .
- ↑ A. Audhya, A. Desai: Proteomics in Caenorhabditis elegans. In: Briefings in functional genomics & proteomics. Volume 7, number 3, May 2008, pp. 205-210, doi: 10.1093 / bfgp / eln014 . PMID 18372286 .
- ↑ S. Mizukami: Development of molecular imaging tools to investigate protein functions by chemical sample design. In: Chemical & pharmaceutical bulletin. Volume 59, Number 12, 2011, pp. 1435-1446, PMID 22130363 .
- ^ A. Ross, W. Kessler, D. Krumme, U. Menge, J. Wissing, J. van den Heuvel, L. Flohé: Optimized fermentation strategy for 13C / 15N recombinant protein labeling in Escherichia coli for NMR structure analysis. In: Journal of biotechnology. Volume 108, Number 1, February 2004, pp. 31-39, PMID 14741767 .
- ↑ J. Li, S. Lin, J. Wang, S. Jia, M. Yang, Z. Hao, X. Zhang, PR Chen: Ligand-free palladium-mediated site-specific protein labeling inside gram-negative bacterial pathogens. In: Journal of the American Chemical Society. Volume 135, number 19, May 2013, pp. 7330-7338, doi: 10.1021 / ja402424j . PMID 23641876 .
- Jump up ↑ Ilya A. Osterman, Alexey V. Ustinov, Denis V. Evdokimov, Vladimir A. Korshun, Petr V. Sergiev, Marina V. Serebryakova, Irina A. Demina, Maria A. Galyamina, Vadim M. Govorun, Olga A. Dontsova : A nascent proteome study combining click chemistry with 2DE Archived from the original on June 30, 2015. Info: The archive link has been inserted automatically and has not yet been checked. Please check the original and archive link according to the instructions and then remove this notice. In: PROTEOMICS . 13, No. 1, January 2013, pp. 17-21. doi : 10.1002 / pmic.201200393 . PMID 23161590 . Retrieved June 27, 2015.
- ↑ C. Uttamapinant, MI Sanchez, DS Liu, JZ Yao, AY Ting: Site-specific protein labeling using PRIME and chelation-assisted click chemistry. In: Nature protocols. Volume 8, number 8, August 2013, pp. 1620–1634, doi: 10.1038 / nprot.2013.096 . PMID 23887180 .
- ^ F. Truong, TH Yoo, TJ Lampo, DA Tirrell: Two-strain, cell-selective protein labeling in mixed bacterial cultures. In: Journal of the American Chemical Society. Volume 134, number 20, May 2012, pp. 8551-8556, doi: 10.1021 / ja3004667 . PMID 22575034 . PMC 3443257 (free full text).
- ↑ ME Ourailidou, JY van der Meer, BJ Baas, M. Jeronimus-Stratingh, AL Gottumukkala, GJ Poelarends, AJ Minnaard, FJ Dekker: Aqueous oxidative heck reaction as a protein-labeling strategy. In: Chembiochem: a European journal of chemical biology. Volume 15, number 2, January 2014, pp. 209–212, doi: 10.1002 / cbic.201300714 . PMID 24376051 .
- ↑ S. Voss, L. Zhao, X. Chen, F. Gerhard, YW Wu: Generation of an intramolecular three-color fluorescence resonance energy transfer probe by site-specific protein labeling. In: Journal of peptide science: an official publication of the European Peptide Society. [electronic publication before going to press] January 2014, doi: 10.1002 / psc.2590 . PMID 24395760 .
- ↑ Z. Yu, Y. Pan, Z. Wang, J. Wang, Q. Lin: Genetically encoded cyclopropene directs rapid, photoclick-chemistry-mediated protein labeling in mammalian cells. In: Angewandte Chemie. Volume 51, number 42, October 2012, pp. 10600-10604, doi: 10.1002 / anie.201205352 . PMID 22997015 . PMC 3517012 (free full text).
- ↑ DP Nguyen, T. Elliott, M. Holt, TW Muir, JW Chin: Genetically encoded 1,2-aminothiols facilitate rapid and site-specific protein labeling via a bio-orthogonal cyanobenzothiazole condensation. In: Journal of the American Chemical Society. Volume 133, number 30, August 2011, pp. 11418-11421, doi: 10.1021 / ja203111c . PMID 21736333 .
- ^ RK Lim, Q. Lin: Photoinducible bioorthogonal chemistry: a spatiotemporally controllable tool to visualize and perturb proteins in live cells. In: Accounts of chemical research. Volume 44, Number 9, September 2011, pp. 828-839, doi: 10.1021 / ar200021p . PMID 21609129 . PMC 3175026 (free full text).
- ^ GC Rudolf, W. Heydenreuter, SA Sieber: Chemical proteomics: ligation and cleavage of protein modifications. In: Current opinion in chemical biology. Volume 17, number 1, February 2013, pp. 110-117, doi: 10.1016 / j.cbpa.2012.11.007 . PMID 23273612 .
- ↑ Y. Takaoka, A. Ojida, I. Hamachi: protein organic chemistry and applications for labeling and engineering in live-cell system. In: Angewandte Chemie. Volume 52, number 15, April 2013, pp. 4088-4106, doi: 10.1002 / anie.201207089 . PMID 23426903 .
- ↑ M. Fernández-Suárez, H. Baruah, L. Martínez-Hernández, KT Xie, JM Baskin, CR Bertozzi, AY Ting: Redirecting lipoic acid ligase for cell surface protein labeling with small-molecule probes. In: Nature Biotechnology . Volume 25, Number 12, December 2007, pp. 1483-1487, doi: 10.1038 / nbt1355 . PMID 18059260 . PMC 2654346 (free full text).
- ^ DW Romanini, VW Cornish: Protein labeling: Playing tag with proteins. In: Nature chemistry. Volume 4, number 4, April 2012, pp. 248-250, doi: 10.1038 / nchem . 1325 . PMID 22437705 .
- ↑ D. Maurel, S. Banala, T. Laroche, K. Johnsson: Photoactivatable and photoconvertible fluorescent probes for protein labeling. In: ACS chemical biology. Volume 5, Number 5, May 2010, pp. 507-516, doi: 10.1021 / cb1000229 . PMID 20218675 .
- ↑ D. Srikun, AE Albers, CI Nam, AT Iavarone, CJ Chang: Organelle-targetable fluorescent probes for imaging hydrogen peroxide in living cells via SNAP-Tag protein labeling. In: Journal of the American Chemical Society. Volume 132, Number 12, March 2010, pp. 4455-4465, doi: 10.1021 / ja100117u . PMID 20201528 . PMC 2850560 (free full text).
- ^ AA Ruggiu, M. Bannwarth, K. Johnsson: Fura-2FF-based calcium indicator for protein labeling. In: Organic & biomolecular chemistry. Volume 8, Number 15, August 2010, pp. 3398-3401, doi: 10.1039 / c000158a . PMID 20556282 .
- ^ C Zhao, LM Hellman, X Zhan, WS Bowman, SW Whiteheart, MG Fried: Hexahistidine-tag-specific optical probes for analyzes of proteins and their interactions . In: Analytical Biochemistry . 399, No. 2, 2010, pp. 237-45. doi : 10.1016 / year from 2009.12.028 . PMID 20036207 . PMC 2832190 (free full text).
- ↑ SR Adams, RE Campbell, LA Gross, BR Martin, GK Walkup, Y. Yao, J. Llopis, Roger Yonchien Tsien : New biarsenical ligands and tetracysteine motifs for protein labeling in vitro and in vivo: synthesis and biological applications. In: Journal of the American Chemical Society. Volume 124, Number 21, May 2002, pp. 6063-6076, PMID 12022841 .
- ↑ T. Machleidt, M. Robers, GT Hanson: Protein labeling with FlAsH and ReAsH. In: Methods in molecular biology. Volume 356, 2007, pp. 209-220, PMID 16988405 .
- ↑ H. Nonaka, SH Fujishima, SH Uchinomiya, A. Ojida, I. Hamachi: FLAG-tag selective covalent protein labeling via a binding-induced acyl-transfer reaction. In: Bioorganic & medicinal chemistry letters. Volume 19, number 23, December 2009, pp. 6696-6699, doi: 10.1016 / j.bmcl.2009.09.122 . PMID 19837586 .
- ↑ GM Eldridge, GA Weiss: Hydrazide reactive peptide tags for site-specific protein labeling. In: Bioconjugate Chemistry . Volume 22, Number 10, October 2011, pp. 2143-2153, doi: 10.1021 / bc200415v . PMID 21905743 . PMC 3199291 (free full text).
- ↑ A. Yoshimura, S. Mizukami, Y. Hori, S. Watanabe, K. Kikuchi: Cell-surface protein labeling with luminescent nanoparticles through biotinylation by using mutant beta-lactamase-tag technology. In: Chembiochem: a European journal of chemical biology. Volume 12, number 7, May 2011, pp. 1031-1034, doi: 10.1002 / cbic.201100021 . PMID 21425232 .
- ↑ C. Uttamapinant, KA White, H. Baruah, S. Thompson, M. Fernández-Suárez, S. Puthenveetil, AY Ting: A fluorophore ligase for site-specific protein labeling inside living cells. In: Proceedings of the National Academy of Sciences . Volume 107, Number 24, June 2010, pp. 10914-10919, doi: 10.1073 / pnas.0914067107 . PMID 20534555 . PMC 2890758 (free full text).
- ^ IV Thiel, G. Volkmann, S. Pietrokovski, HD Mootz: An atypical naturally split intein engineered for highly efficient protein labeling. In: Angewandte Chemie. Volume 53, number 5, January 2014, pp. 1306-1310, doi: 10.1002 / anie.201307969 . PMID 24382817 .
- ^ I. Ghosh, N. Considine, E. Maunus, L. Sun, A. Zhang, J. Buswell, TC Evans, MQ Xu: Site-specific protein labeling by internal-mediated protein ligation. In: Methods in molecular biology. Volume 705, 2011, pp. 87-107, doi : 10.1007 / 978-1-61737-967-3_6 . PMID 21125382 .
- ↑ JY Yang, WY Yang: Site-specific two-color protein labeling for FRET studies using split inteins. In: Journal of the American Chemical Society. Volume 131, Number 33, August 2009, pp. 11644-11645, doi: 10.1021 / ja9030215 . PMID 19645470 .
- ↑ Y. Du, CE Hendrick, KS Frye, LR Comstock: Fluorescent DNA labeling by N-mustard analogues of S-adenosyl-L-methionine. In: Chembiochem: a European journal of chemical biology. Volume 13, number 15, October 2012, pp. 2225–2233, doi: 10.1002 / cbic.201200438 . PMID 22961989 .
- ↑ SE Walker, J. Lorsch: Sanger dideoxy sequencing of DNA. In: Methods in enzymology. Volume 529, 2013, pp. 171-184, doi: 10.1016 / B978-0-12-418687-3.00014-8 . PMID 24011045 .
- ^ R. Redon, T. Fitzgerald, NP Carter: Comparative genomic hybridization: DNA labeling, hybridization and detection. In: Methods in molecular biology. Volume 529, 2009, pp. 267-278, doi : 10.1007 / 978-1-59745-538-1_17 . PMID 19381974 . PMC 2867219 (free full text).
- ↑ H. Zohar, SJ Muller: Labeling DNA for single-molecule experiments: methods of labeling internal specific sequences on double-stranded DNA. In: Nanoscale. Volume 3, Number 8, August 2011, pp. 3027-3039, doi: 10.1039 / c1nr10280j . PMID 21734993 . PMC 3322637 (free full text).
- ↑ E. Paredes, M. Evans, SR Das: RNA labeling, conjugation and ligation. In: Methods. Volume 54, Number 2, June 2011, pp. 251-259, doi: 10.1016 / j.ymeth.2011.02.008 . PMID 21354310 .
- ↑ DV Bann, LJ Parent: Application of live-cell RNA imaging techniques to the study of retroviral RNA trafficking. In: Viruses. Volume 4, Number 6, June 2012, pp. 963-979, doi: 10.3390 / v4060963 . PMID 22816035 . PMC 3397357 (free full text).
- ↑ JS Kaddis, DH Wai, J. Bowers, N. Hartmann, L. Baeriswyl, S. Bajaj, MJ Anderson, RC Getts, TJ Triche: Influence of RNA labeling on expression profiling of microRNAs. In: The Journal of Molecular Diagnostics , Volume 14, Number 1, January 2012, pp. 12-21, doi: 10.1016 / j.jmoldx.2011.08.005 . PMID 22074760 . PMC 3338349 (free full text).
- ↑ K. Cole, V. Truong, D. Barone, G. McGall: Direct labeling of RNA with multiple biotins allows sensitive expression profiling of acute leukemia class predictor genes. In: Nucleic Acids Research , Volume 32, Number 11, 2004, p. E86, doi: 10.1093 / nar / gnh085 . PMID 15205470 . PMC 443553 (free full text).
- ↑ V. Olieric, U. Rieder, K. Lang, A. Serganov, C. Schulze-Briese, R. Micura, P. Dumas, E. Ennifar: A fast selenium derivatization strategy for crystallization and phasing of RNA structures. In: RNA. Volume 15, Number 4, April 2009, pp. 707-715, doi: 10.1261 / rna.1499309 . PMID 19228585 . PMC 2661828 (free full text).
- ↑ SC Walker, FH Scott, C. Srisawat, DR Engelke: RNA affinity tags for the rapid purification and investigation of RNAs and RNA-protein complexes. In: Methods in molecular biology. Volume 488, 2008, pp. 23-40, doi : 10.1007 / 978-1-60327-475-3_3 . PMID 18982282 . PMC 2807123 (free full text).
- ↑ JS Paige, KY Wu, SR Jaffrey: RNA mimics of green fluorescent protein. In: Science. Volume 333, Number 6042, July 2011, pp. 642–646, doi: 10.1126 / science.1207339 . PMID 21798953 . PMC 3314379 (free full text).
- ^ RM Martin, J. Rino, C. Carvalho, T. Kirchhausen, M. Carmo-Fonseca: Live-cell visualization of pre-mRNA splicing with single-molecule sensitivity. In: Cell reports. Volume 4, number 6, September 2013, pp. 1144–1155, doi: 10.1016 / j.celrep.2013.08.013 . PMID 24035393 . PMC 3805459 (free full text).
- ^ JA Prescher, CR Bertozzi: Chemical technologies for probing glycans. In: Cell. Volume 126, Number 5, September 2006, pp. 851-854, doi: 10.1016 / j.cell.2006.08.017 . PMID 16959565 .
- ↑ XL Sun, W. Cui, C. Haller, EL Chaikof: Site-specific multivalent carbohydrate labeling of quantum dots and magnetic beads. In: Chembiochem: a European journal of chemical biology. Volume 5, Number 11, November 2004, pp. 1593-1596, doi: 10.1002 / cbic.200400137 . PMID 15515080 .