Sortase
Sortases form a family of membrane-associated bacterial enzymes and are assigned to the class of transpeptidases .
properties
The biological function of the sortases is to covalently anchor secreted proteins in the peptidoglycan layer of gram-positive bacteria. In addition, there are sortases that are responsible for building pilus structures from individually expressed protein subunits. Therefore, sortases are to a large extent responsible for the structure of the cell surface of gram-positive bacteria.
Since there are many virulence factors on the bacterial surface , sortases are being discussed as a possible therapeutic target in the treatment of infectious diseases .
The sortase reaction mechanism can be used in vitro to link proteins (including other molecules) covalently and site-specifically, which is why the enzymes are also used in biotechnology. It is advantageous here that functional sortases can be expressed heterologously (e.g. in E. coli ), which enables production on a larger scale with relatively little effort.
Sortases require divalent cations (e.g. Ca 2+ ) as cofactors .
Occurrence and classification
Sortases are found in almost all gram-positive bacteria, but can also be found in isolated cases in gram-negative and archaea . To date, over 800 genes in more than 260 different species have been identified that encode sortase-related proteins. About 60% of these proteins can be divided into a total of six different groups (A – F).
The individual groups are differentiated according to their function and their substrate specificity . While groups A and B mainly have the function of anchoring proteins in the peptidoglycan layer of the bacteria, sortases of groups C and D are involved in the construction of pilus structures. Furthermore, the group D sortases are frequently found in bacteria of the genus Bacillus , in which they are associated with the formation of endospores . The functions of sortases E and F are not yet fully understood.
As a rule, gram-positive bacteria have more than one type of sortase.
Reaction mechanism
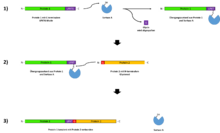

Basically, sortases are able to covalently link two peptide residues with one another. To do this, they need a specific recognition motif, which often consists of five amino acids . Sortases can split these recognition motifs within a protein and then reassemble them with another ligand. The identification motifs can differ, depending on the type of sortase and organism.
| organism | Sortase type | Recognition motif |
|---|---|---|
| Staphylococcus aureus | Sortase A | LPXTG |
| Staphylococcus aureus | Sortase B | NPQTN |
| Streptococcus pneumoniae | Sortase B | YPRTG |
In vitro reaction
In the following section, the sortase reaction mechanism is illustrated using the well-characterized sortase A from Staphylococcus aureus . The recognition motif of this sortase is LPXTG (leucine-proline-Xaa-threonine-glycine). X stands for any amino acid.
The reaction takes place in two steps. First, Sortase A binds to the protein / peptide. The covalent bond takes place between the thiol residue of a cysteine in the active center of sortase A and the threonine within the LPXTG recognition sequence. As a result, the C-terminal residue of the protein, including the glycine , is split off. A transition state arises in which sortase A is covalently linked to the protein residue. In the second step, this transition state meets a protein / peptide with a free, N-terminal, glycine residue. The two proteins are covalently linked to each other by sortase A, which re-establishes the original peptide bond (the LPXTG motif).
In vivo function of sortase A from Staphylococcus aureus
Proteins that are intended to be incorporated into the cell wall of S. aureus have several functional subunits. This includes an N -terminal signal peptide (SP), which is responsible for ensuring that the protein is secreted from the cytoplasm - through the membrane - into the extracellular space. The signal peptide is removed in the process.
The secreted protein is then attached to the cell membrane by a C -terminal hydrophobic region (HR) . The now membrane-associated protein carries the LPXTG recognition motif to which the membrane-associated sortase binds. By binding the sortase to the LPXTG motif, the hydrophobic region is split off and a transitional state is created in which the sortase is linked to the protein.
Through the reaction of the transition state with the free pentaglycine residue of lipid II (a precursor of the peptidoglycan building blocks), the secreted protein is covalently anchored to the cell wall building block (lipid II). As the cell wall synthesis continues, the protein becomes part of the peptidoglycan layer.
Applications
Sortases are used in biotechnology to connect molecules with one another and thus spatially couple their function. Not only proteins can be linked, but also lipids and other molecules. The prerequisite is that the recognition sequences for the sortase can be attached to the molecules. The motifs must also be correctly oriented and spatially accessible for the sortase.
Proteins or other molecules can be marked for analytical purposes with the help of sortases . For example, antibodies or receptor ligands can be labeled with a fluorescent dye in order to examine their binding properties.
With the help of the sortase reaction mechanism, therapeutic molecules can be PEGylated . By combining a therapeutic protein with polyethylene glycol (PEG), the breakdown of the protein in the body can be slowed down. With the longer biological half-life of the therapeutic protein, the pharmacodynamic properties are improved.
By connecting the C and N terminus of a protein, a ring closure is achieved. This inhibits the breakdown of therapeutic proteins by proteases . Like PEGylation, this leads to a longer biological half-life of the protein.
The use of several sortases with different recognition motifs allows the synthesis of a complex from several ligands. The orientation and sequence of the individual ligands can be controlled.
Advantages of using sortases
Sortases are enzymes and function under physiological conditions. Therefore, in contrast to some chemical processes, sortases can be used without any damage to the substrates (e.g. due to temperature, pH value or salt concentration) being expected.
The reaction of the sortases is always specific, which means that the ligation of the substrates always takes place in the same place. In chemical synthesis, on the other hand, the substrates can sometimes be linked at different points.
Disadvantages of using sortases
Sortases catalyze the back and forth reaction. This means that they catalyze both the linking of the substrates and the loosening of the bond. The reason for this is that the linkage restores the sortase's recognition motif. As a result, a reaction equilibrium between the substrates and the product is established after a certain period of time . The product yield of the sortase reaction depends on the properties of the substrates and the conversion is usually not complete.
The purification of the product from the reaction batch can also be problematic. However, this also depends heavily on the substrates used and therefore cannot be assessed across the board.
Sortases as a target of therapeutic applications
Sortases are to a large extent responsible for the architecture of the cell wall of gram-positive bacteria. The surface structures that are built up by sortases also include many virulence factors which z. B. are necessary for the metabolism of bacteria or for adhesion to human tissue. Studies have shown that pathogens whose sortases have been genetically inactivated are no longer able to trigger infections. From this one concludes that a medicinal inhibition of the sortases could represent an antibiotic therapy.
Individual evidence
- ↑ SK Mazmanian, G. Liu, ER Jensen, E. Lenoy, O. Schneewind: Staphylococcus aureus sortase mutants defective in the display of surface proteins and in the pathogenesis of animal infections. In: Proceedings of the National Academy of Sciences . Volume 97, number 10, May 2000, ISSN 0027-8424 , pp. 5510-5515, doi: 10.1073 / pnas.080520697 , PMID 10805806 , PMC 25859 (free full text).
- ↑ a b Ki-Bong Oh et al. : Inhibition of sortase-mediated Staphylococcus aureus adhesion to fibronectin via fibronectin-binding protein by sortase inhibitors. In: Applied Microbiology and Biotechnology . 70, No. 1 2006, pp. 102-106.
- ↑ a b c Markus Hilleringmann et al. : Molecular architecture of Streptococcus pneumoniae TIGR4 pili. In: The EMBO Journal . 28, No. 24 2009, pp. 3921-3930.
- ↑ a b Luciano A. Marraffini, Andrea C. Dedent and Olaf Schneewind: Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. In: Microbiology and molecular biology reviews: MMBR. 70, No. 1 2006, pp. 192-221.
- ↑ Stella Cascioferro, Makrina Totsika and Domenico Schillaci: Sortase A: an ideal target for anti-virulence drug development. In: Microbial pathogenesis. 77 2014, pp. 105–112.
- ↑ CS Theile, MD Witte, AE Blom, L. Kundrat, HL Ploegh, CP Guimaraes: Site-specific N-terminal labeling of proteins using sortase-mediated reactions. In: Nature protocols. Volume 8, number 9, September 2013, ISSN 1750-2799 , pp. 1800-1807, doi: 10.1038 / nprot.2013.102 , PMID 23989674 , PMC 3941705 (free full text).
- ↑ U. Ilangovan et al. : Structure of sortase, the transpeptidase that anchors proteins to the cell wall of Staphylococcus aureus. In: Proceedings of the National Academy of Sciences . 98, No. 11 2001, pp. 6056-6061.
- ↑ a b M. J. Pallen et al. : An embarrassment of sortases - a richness of substrates? In: Trends in microbiology. 9, No. 3 2001, pp. 97-102.
- ↑ a b Thomas Spirig, Ethan M. Weiner and Robert T. Clubb: Sortase enzymes in Gram-positive bacteria. In: Molecular microbiology. 82, No. 5 2011, pp. 1044-1059.
- ↑ Jonathan M. Budzik, Luciano A. Marraffini and Olaf Schneewind: Assembly of pili on the surface of Bacillus cereus vegetative cells. In: Molecular microbiology. 66, No. 2 2007, pp. 495-510.
- ↑ a b Gavin K. Paterson and Timothy J. Mitchell: The biology of Gram-positive sortase enzymes. In: Trends in microbiology. 12, No. 2 2004, pp. 89-95.
- ↑ John M. Antos et al. : Lipid modification of proteins through sortase-catalyzed transpeptidation. In: Journal of the American Chemical Society . 130, No. 48 2008, pp. 16338-16343.
- ↑ Maximilian W. Popp et al. : Sortagging: a versatile method for protein labeling. In: Nature Chemical Biology . 3, No. 11 2007, pp. 707-708
- ↑ a b Maximilian W. Popp et al. : Sortase-catalyzed transformations that improve the properties of cytokines. In: Proceedings of the National Academy of Sciences . 108, No. 8 2011, pp. 3169-3174.
- ↑ John M. Antos et al. : Site-specific N- and C-terminal labeling of a single polypeptide using sortases of different specificity. In: Journal of the American Chemical Society. 131, No. 31 2009, pp. 10800-10801.
- ↑ Sarkis K. Mazmanian et al. : An iron-regulated sortase anchors a class of surface protein during Staphylococcus aureus pathogenesis. In: Proceedings of the National Academy of Sciences . 99, No. 4 2002, pp. 2293-2298.
- ↑ Anthony W. Maresso and Olaf Schneewind: Sortase as a target of anti-infective therapy. In: Pharmacological Reviews . 60, No. 1 2008, pp. 128-141.


