In situ hybridization

In situ hybridization ( ISH , including in situ hybridization ) is a molecular biological method to detect nucleic acids ( RNA or DNA ) in tissues , individual cells or metaphase - chromosomes . An artificially produced probe from a nucleic acid is used, which binds to the nucleic acid to be detected via base pairing ( hybridization ).
Widespread have the fluorescence in situ hybridization (FISH) for detection of DNA or RNA (RNA-FISH) in cell nuclei of individual cells or metaphase chromosomes as well as the investigation of the distribution of mRNA in whole embryos, sections or tissues with coloring (chromogenic) molecules ( FISH test ).
Other terms used are genomic in-situ hybridization (GISH), in which total genomic DNA is used as a probe , and chromosomal in-situ suppression hybridization (CISS hybridization), with which whole chromosomes can be marked.
history
In situ hybridization was developed in the late 1960s by the American biologists Mary Lou Pardue and Joe Gall . They used radioactively labeled probes ( tracers ), the specimens then had to be placed on an X-ray film. In addition to the general problems associated with radioactivity, the spatial resolution was poor compared to current techniques. Therefore, probes that were covalently bound to the marker molecules later prevailed (see below).
functionality
In situ hybridization is based on the pairing of complementary bases on two nucleic acid single strands. One of the two strands comes from a previously produced and marked probe , the other is in the preparation and is to be detected. The first steps are therefore preparation of the specimen and preparation of the probe. The probe usually consists of DNA, as it is more stable than RNA and is therefore easier to handle in the laboratory. DNA can also be multiplied more easily with today's methods. The probe can be labeled indirectly with haptens (e.g. digoxigenin , biotin or 2,4-dinitrophenol ) or, with FISH, directly with fluorescent molecules such as FITC or Cy3. Frequently used methods for labeling the probes are nick translation , PCR and in vitro transcription .
Since DNA normally exists as a double strand, it must first be separated (also: melted or denatured). Denaturation can occur either by shifting the pH value (both acidic and alkaline) or by heat. In the case of heat denaturation, the melting point is usually lowered by adding formamide , which, due to its strong polarity, weakens the hydrogen bonds between the individual DNA strands. This means that melting can be achieved at temperatures of around 70–75 ° C, which means that the structure of the chromosomes is less damaged. Furthermore, the DNA can also be stretched and aligned by molecular combining .
The actual hybridization takes between one hour and several days, depending on the probe material and target sequence. Hybrid molecules from the nucleic acids of the preparation and the probe are then present in the preparation. Unbound or unspecifically bound probe molecules are washed out and bound probe molecules can be detected. In the case of indirect labeling, this is done via antibody staining or, in the case of biotin, with avidin . The antibodies (or the avidin) are in turn bound to fluorophores (in FISH) or to enzymes which form dyes from the chromogenic substrates to be added. Both are then evaluated microscopically.
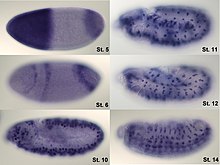
Example of RNA in situ hybridization at different stages of Drosophila melanogaster embryos. The digoxigenin- labeled antisense RNA probe is directed against the mRNA of the transcription factor hunchback . It was made visible by a coloring reaction with alkaline phosphatase . It can be clearly seen that the expression pattern of the gene changes in the various stages of development (St. stands for stage).
|

The activity of a gene in a mouse embryo of the embryonic stage E14.5 (after 14.5 days out of a total of 19.5 days of gestation). Using in situ hybridization, the presence of the mRNA and thus the activity of the cannabinoid receptor type 1 (Cnr1) gene was made visible in a 25 micrometer thin frozen section , which can be seen as a dark blue color in various areas of the brain and in the spinal cord.
|
Detection of mRNA with coloring enzymes
In this application, mainly found digoxigenin -labeled RNA using. Digoxigenin can be recognized with the aid of an antibody that is coupled to an enzyme , for example . The enzyme, usually alkaline phosphatase or peroxidase , can then convert a dye through the addition of reagents, which remains covalently bound in the tissue and is therefore not distributed by diffusion . When mRNA is detected in tissues, only those cells are stained ( hybridized ) in which a gene to be investigated is active: only here is the corresponding mRNA available. This method is particularly used in developmental biology . Here it is of particular interest to track the activity of a gene in situ, for example during embryogenesis . The embryonic or adult tissue must first be fixed for the staining; activity cannot therefore be tracked in real time , it is just a snapshot of the state the tissue was in when it was fixed.
procedure
The tissue to be stained - for example embryos from various model organisms such as Arabidopsis thaliana , Drosophila melanogaster , Xenopus laevis , Mus musculus , Danio rerio - is fixed with the help of solutions containing formaldehyde . It is then transferred to a formamide-containing buffer and the labeled probe is added. The hybridization time depends on the size of the embryo and takes at least a few hours. During this time, the probe can diffuse through the tissue and binds wherever complementary sequences are found in the mRNA. This is followed by a few washing steps in which excess, unbound probes are washed out, and finally the staining, which can then be precisely analyzed and documented with a microscope .
In addition to total specimens, for example from fly embryos (left figure), in situ hybridization can be carried out on thin tissue sections. In this way, larger specimens such as human tissue or complete mouse embryos can also be examined (right figure).
Fluorescence in situ hybridization
With fluorescence in situ hybridization (FISH), the probe is detected with the aid of a fluorescent dye. This enables the simultaneous detection of several structures through the use of different fluorescent dyes, for example the detection of a chromosome with two genes lying on it. Up to seven different dyes have already been successfully used on chromosome preparations and on cell nuclei.
Metaphase chromosomes (blue) with evidence of a translocation through the use of two gene-specific probes (green and red), see Philadelphia chromosome
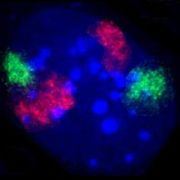
Cell nucleus of a mouse fibroblast . The territories of chromosomes 2 (red) and 9 (green) were stained by fluorescence in situ hybridization . DNA counterstaining in blue. ( This can also be found in other cell types of the mouse a blackboard with further examples.)
Gene-rich and gene-poor regions in human chromosomes. On the metaphase chromosomes of a human female lymphocyte , the Alu sequences were marked (green) by fluorescence in situ hybridization , which are particularly common in gene-rich chromosome sections. The DNA is colored red for the visibility of gene-poor regions.
application areas
The possibility of determining the presence or number of parts of the genome comparatively easily in a fluorescence microscope is used for clinical diagnostics in various disciplines.
For the detection of hereditary diseases , preparations of chromosomes in the metaphase are usually used, which have been produced from cells of patients that have been reproduced in the laboratory or, for prenatal diagnosis , from cells of the unborn child (see also FISH test ). Probes for chromosome 21, for example, are then hybridized to this in order to test whether it is present three times instead of twice and thus whether the cause of Down's syndrome is present. Whether certain chromosomal regions are missing, such as in cat cry syndrome , can also be determined with probes for such regions. In principle, such examinations can also be carried out on cell nuclei, i.e. without metaphase preparations having to be made. However, this has the disadvantage that not every nucleus delivers a correct result: If two chromosomes to be detected happen to be next to each other in the nucleus, then both cannot be recognized as two signals. Therefore, many nuclei have to be counted and a comparison with known normal nuclei has to be carried out in order to be able to make a reliable statement. The advantage of this method lies in the higher speed, since a multiplication of the cells in the laboratory is not necessary or only to a lesser extent. FISH is also used to characterize marker chromosomes .
When examining the genome of tumor cells , FISH is used to detect chromosome aberrations . For example, metaphase preparations can be examined with FISH in order to detect chromosomal alterations that can not be diagnosed by G-banding . To do this, probes for different human chromosomes can be tried in sequence. Multi-color FISH also makes it possible to detect all chromosomes in different color combinations, so that all changes between different chromosomes can be recognized in one experiment. However, the methods required for this are demanding and require special microscopic equipment. Some chromosomal modifications occur more frequently in certain types of tumors, for example the Philadelphia chromosome in some leukemias . In non-Hodgkin lymphoma , different subgroups have different typical genetic changes. The presence of such changes can be specifically tested with FISH.
literature
- AR Leitch, T. Schwarzacher, D. Jackson, IJ Leitch: In situ hybridization. Spectrum Academic Publishing House, Heidelberg / Berlin 1994.
- JM Levsky, R. Singer: Fluorescence in situ hybridization: past, present and future. In: Journal of Cell Science. 116, No. 14, pp. 2833-2838, PMID 12808017 , doi: 10.1242 / jcs.00633 .
Web links
- GenePaint.org (database of gene expression patterns in the mouse embryo examined with in situ hybridization)
Individual evidence
- ↑ Joseph G. Gall, Mary Lou Pardue: Formation and detection of RNA-DNA hybrid molecules in cytological preparations. In: Proc. Natl. Acad. Sci. USA 63, No. 2, 1969, pp. 378-383, PMID 4895535 , PMC 223575 (free full text).
- ^ Mary Lou Pardue, Joseph G. Gall: Molecular hybridization of radioactive DNA to the DNA of cytological preparations. In: Proc. Natl. Acad. Sci. USA 64, No. 2, 1969, pp. 600-604, PMID 5261036 , PMC 223386 (free full text).
- ↑ Andreas Bolzer, Gregor Kreth, Irina Solovei, Daniela Koehler, Kaan Saracoglu, Christine Fauth, Stefan Müller, Roland Eils, Christoph Cremer , Michael R. Speicher, Thomas Cremer: Three-dimensional maps of all chromosome positions demonstrate a probabilistic order in human male fibroblast nuclei and prometaphase rosettes. In: PLoS Biology . 3, No. 5, 2005, p. E157, PMID 15839726 , doi: 10.1371 / journal.pbio.0030157 .
- ↑ L. Brecevic, S. Michel, H. Starke, K. Müller, N. Kosyakova, K. Mrasek, A. Weise, T. Liehr: Multicolor FISH used for the characterization of small supernumerary marker chromosomes (sSMC) in commercially available immortalized cell lines. In: Cytogenetic & Genome Research. 114, No. 3,4, 2006, pp. 319-324, doi: 10.1159 / 000094220 .
- ↑ Michael R. Speicher, Stephen Gwyn Ballard, David C. Ward: Karyotyping human chromosomes by combinatorial multi-fluor FISH. In: Nature Genetics . 12, 1996, pp. 368-375, doi: 10.1038 / ng0496-368 .
- ↑ E. Schröck, S. du Manoir, T. Veldman, B. Schoell, J. Wienberg, MA Ferguson-Smith, Y. Ning, DH Ledbetter, I. Bar-Am, D. Soenksen, Y. Garini, T. Ried: Multicolor Spectral Karyotyping of Human Chromosomes. In: Science . 273, No. 5274, 1996, pp. 494-497, doi: 10.1126 / science.273.5274.494 .