Philadelphia chromosome

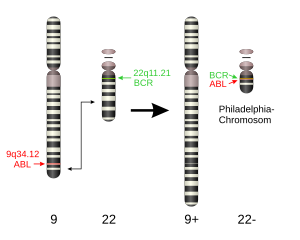
The Philadelphia chromosome (obsolete Ph 1 ) is a shortened chromosome 22 that can be found in some human leukemias . It is caused by a chromosome translocation between chromosomes 9 and 22 . The cytogenetic notation for the translocation is: t (9; 22) (q34; q11).
The Philadelphia chromosome was first described in 1960 by Peter Nowell and David Hungerford in Philadelphia in leukemia cells from a patient with chronic myeloid leukemia (CML) and was named from the place of discovery. It was the first identified chromosomal change that could be linked to the development of cancer . The change is detectable in more than 95 percent of CML patients. It was later discovered that it can also be found in some patients with acute lymphoblastic leukemia (ALL) (around 4% of cases in children and 25% in adults), very rarely also in acute myeloid leukemia (AML; in less than one percent of cases).
Origin and consequences
Translocation
The chromosome change takes place in the bone marrow in a stem cell of the blood . Chromosome 9 breaks in area q34.1 (q denotes the long chromosome arm, 34.1 the position on this) and chromosome 22 on q11.2. The breakpoint is on both chromosomes in the area of genes , the ABL gene (or ABL1 ; for Abelson Murine Leukemia Viral Oncogene Homolog 1 ) on chromosome 9 and the BCR gene ("breakpoint cluster region"; named because of the frequent breaks in this gene) on chromosome 22.
During the translocation, the 5 'part of the BCR gene is linked to the 3' part of the ABL gene. There are different possible breakpoints in the BCR gene, but only one breakpoint in the ABL gene, so that fusion genes of different sizes arise, in which the portion of the ABL gene is always the same, but the size of the BCR portion varies. This leads to the formation of the fusion genes BCR-ABL on chromosome 22 and ABL-BCR on chromosome 9, which is now called 9q + in its extended form. In the so-called Philadelphia-positive leukemias (mostly CML), the chromosome translocation is visible on a cytogenetic examination as a shortened chromosome 22, the Philadelphia chromosome described here. The gene can also be identified using a specific polymerase chain reaction .
In most cases, the cause of this chromosome change is not known or cannot be determined. In very few cases, a radiation accident ( ionizing radiation ) or benzene can be considered as the cause .
Gene product
The mutual (reciprocal) translocation t (9; 22) (q34; q11) as well as the new fusion genes result in an altered gene product in both chromosomes. The newly created BCR-ABL gene on chromosome 22 is transcribed in the cell , creating the new protein BCR-ABL gene product , a fusion protein . The translation of the resulting mRNA leads to the synthesis of the modified protein. The enzyme originally transcribed by the ABL gene is a tyrosine kinase and plays an important role in cellular growth regulation. The fusion protein consists of the amino end of the BCR protein and the carboxy end of the ABL protein, which contains a kinase domain . As a result, the tyrosine kinase activity is permanently activated under the influence of the BCR region and the affected cell multiplies in an uncontrolled manner (deficient apoptosis ). This development turns the cell into a tumor cell .
The specific mechanism by which the new fusion gene leads to uncontrolled proliferation has not yet been fully elucidated. The ABL gene normally has two introductory exons 1a and 1b, which can be used alternatively for transcription. The selection takes place during splicing with exon 2, which has a so-called splice acceptor site and allows either 1a or 1b to dock. During the translocation, exon 1 is exchanged for the fragment of the BCR , which is also accepted by exon 2 and added to the gene and thus transcribed together with exons 2 to 11 of the ABL .
Oncogenic effects
See main articles Chronic Myeloid Leukemia and Acute Lymphocytic Leukemia
Due to the changed tyrosine kinase activity of the ABL gene under the influence of the BCR region, the affected cell multiplies in an uncontrolled manner and becomes a tumor cell. As a pluripotent stem cell, it produces different cell types that also contain the changed chromosome. All cells derived from this stem cell contain the changed pair of chromosomes 9 and 22 and thus the Philadelphia chromosome. A pathological effect, however, only occurs in the leukemic white blood cells .
Since ABL , like other versions of the ABL gene (for v-ABL see below), becomes an oncogene by changing the initial sequence , it is called a proto-oncogene . Similar to other translocations leading to tumor diseases, an oncogene is created here by fusing two normal genes.
The fusion protein binds to various other proteins, including the kinase regulator protein CRK ( CT10 regulator of kinase ), phosphatidylinsitol 3'-kinase and GRB-2 / SOS-1. By binding to the latter, the activation of the RAS gene, which plays a central role in controlling cell growth and proliferation, is increased. RAS mutations, in turn, are considered to be the central triggers of various tumors and could also play a central role in the oncogenic effect of the BCR-ABL gene.
Variations and additional chromosome changes
In detail, the translocations that lead to chronic myeloid leukemia (CML) and acute lymphoblastic leukemia (ALL) differ in the position of the break in the BCR gene of chromosome 22 and thus also in the length of the later BCR-ABL- Gene product. The break in chromosome 9 is always in the same intron . The breakpoint in chromosome 22, however, varies. So far, three breakpoints have been described in the BCR region. They are called m-BCR (minor), M-BCR (major) and µ-BCR (micro). The m-BCR break point is the furthest 5 '. The resulting fusion protein is therefore the smallest at around 190 kDa. The breaking point M-bcr lies further 3 'and leads to a fusion protein with a size of 210 kDa. The breakpoint µ-BCR is the furthest 3 'and leads to the largest BCR-ABL fusion protein with 230 kDa.
Fusion genes, leukemias and cytogenetic status
Three different BCR-abl fusion genes are thus described in the literature. The Ph chromosome occurs not only in CML, but also in around 20 percent of the examined cases in ALL in adults, in five percent of the examined cases in ALL in children and in around two percent of the cases in AML. In addition, some of the patients examined with CML or an adult form of ALL do not have a Ph chromosome, but a fusion gene can be detected. There are also a small number of patients with CML who neither have a Ph chromosome nor express a fusion gene.
Chronic myeloid leukemia
90 to 95 percent of patients with chronic myeloid leukemia (CML) have a Ph chromosome. Over 99 percent of all patients with CML express the 210 kD fusion protein. Around five percent of patients with CML are Ph-negative, of which almost 100 percent of the cases examined also express the 210 kD fusion product. Thus, in CML , the so-called major BCR region located roughly in the middle of the BCR gene is usually . affected, which makes up about 5.8 kb of the gene , which is over 90 kb in size. The resulting gene product has a mass of 210 kDa (P210) compared to the mass of 145 kDa of the original ABL protein, where 140 kDa are accounted for by ABL and 70 kDa by BCR .
Acute lymphoblastic leukemia in adults
A Ph chromosome can be found in about 20 percent of adult patients with ALL using cytogenetic examination methods. The bcr gene breaks in 20-50 percent of cases in the first intron ( minor bcr region ) and the gene product of the fusion gene is only 185 kDa long, of which 45 kDa are due to BCR . The M-BCR breakpoint is affected in 50–80 percent of cases. This leads to the 210 kD fusion protein, which is also mostly found in CML. About 10 percent of adult patients with ALL are Ph negative, but express a BCR-abl fusion gene. In these rare cases, the 190 kDa and 210 kDa forms are found about equally often.
Acute lymphocytic leukemia in childhood
In ALL of the child , a Ph chromosome is found in five percent of the cases examined. These children with a Ph-positive ALL express the 210 kDa fusion protein in 10 percent of the cases examined and the 190 kDa protein in 90 percent. The constellation of a ph-negative child ALL with evidence of the Bcr-Abl fusion protein is not known.
Acute myeloid leukemia
About two percent of the examined cases of patients with AML show a Ph chromosome cytogenetically. In these rare cases, the fusion proteins p210 and p190 occur roughly equally. In very rare cases, one finds patients with Ph-negative AML who express a Bcr-Abl fusion gene.
Other findings
Patients who have a Philadelphia chromosome often have other changed and increased chromosomes in the affected cells, so they develop so-called somatic aneuploidies . In clinical studies of 67 CML patients, an additional Philadelphia chromosome was found in almost 50 percent of those examined (33) and a trisomy of the long arm of chromosome 17 in 28 . In addition to these two forms, other aneuploidies of the cells occurred.
Comparable oncogenes
The effect of the Abelson's mouse leukemia virus , a retrovirus that causes leukemia on B lymphocytes in mice, is comparable to the oncogenic effect of the gene change in the Philadelphia chromosome . Here, too, an ABL gene ( v-ABL ) is changed by a further gene, in this case the GAG gene of the virus, and stimulated to an increased tyrosine kinase activity. Due to the similarity of the genetic modification, the correspondingly diseased mice are used as model organisms for the development of preparations against the oncogenic effects of the chromosome modification in pharmaceutical research as well as for basic research.
Research history
The Philadelphia chromosome was described in 1960 by Peter Nowell of the University of Pennsylvania School of Medicine and David Hungerford of the Fox Chase Cancer Center's Institute for Cancer Research as the first constant chromosomal change in tumor cells in patients with chronic myeloid leukemia . They found a very short chromosome, which they believed to be the Y chromosome , in the blood samples of two patients. Later it turned out that it was the shortened chromosome 22, which was named after the place of its discovery as the Philadelphia chromosome (abbreviated Ph 1 ). In a recent report, PC Nowell shares his personal memories of discovering the Philadelphia chromosome.
In 1972 Janet Rowley was able to show that this chromosome is created by an exchange of genetic material between the long arms of chromosome 9 and chromosome 22.
In 1983 and 1984 it was discovered that there are two genes at the chromosome breakpoints ( ABL and BCR ), which are fused together through chromosome translocation.
Pharmacological research subsequently tried to block the oncogenic effects of the modified gene product. With the help of imatinib , an inhibitor of BCR-ABL tyrosine kinase developed in the 1980s, it is now possible to achieve longer-lasting remissions in CML .
Sources and further information
Individual evidence
- ↑ PC Nowell, DA Hungerford: Chromosome studies on normal and leukemic human leukocytes. In: J Natl Cancer Inst . 1960; 25, pp. 85-109. PMID 14427847
- ↑ Czerwenka et al. 2003, p. 170.
- ↑ Bain 1999, p. 83.
- ↑ Miller & Therman 2001, p. 408.
- ↑ R. Kurzrock et al.: Philadelphia Chromosome-positive leukemias: from basic mechanisms to molecular therapeutics. In: Ann Intern Med . 2003 ; 138, pp. 819-830. PMID 12755554 . (Review, Free Full Text).
- ↑ Czerwenka et al. 2003, pp. 170–171.
- ↑ R. Kurzrock et al .: BCR rearrangement-negative chronic myelogenous leukemia revisited. In: J Clin Oncol . 2001 Jun 1; 19 (11), pp. 2915-2926. PMID 11387365
- ^ R. Kurzrock et al.: Philadelphia chromosome-negative chronic myelogenous leukemia without breakpoint cluster region rearrangement: a chronic myeloid leukemia with a distinct clinical course. In: Blood . 1990 Jan 15; 75 (2), pp. 445-452. PMID 2403827
- ^ R. Kurzrock et al .: Rearrangement in the breakpoint cluster region and the clinical course in Philadelphia-negative chronic myelogenous leukemia. In: Ann Intern Med. 1986 Nov; 105 (5), pp. 673-679. PMID 3094418
- ↑ R. Kurzrock et al .: A novel c-abl protein product in Philadelphia-positive acute lymphoblastic leukaemia. In: Nature . 1987 Feb 12-18; 325 (6105), pp 631-635. PMID 3543692
- ^ R. Kurzrock et al.: Molecular analysis of chromosome 22 breakpoints in adult Philadelphia-positive acute lymphoblastic leukaemia. In: Br J Haematol . 1987 Sep; 67 (1), pp. 55-59. PMID 3478080
- ↑ J. Erikson et al: Heterogeneity of chromosome 22 breakpoint in Philadelphia-positive (Ph +) acute lymphocytic leukemia. In: Proc Natl Acad Sci USA . 1986 Mar; 83 (6), pp. 1807-1811. PMID 3513189
- ^ R. Kurzrock et al .: Expression of c-abl in Philadelphia-positive acute myelogenous leukemia. In: Blood. 1987 Nov; 70 (5), pp. 1584-1588. PMID 3311207 .
- ^ David T. Suzuki, Anthony JF Griffiths, Jeffrey H. Miller, Richard C. Lewontin: Genetics. 1st edition. VCH Verlagsgesellschaft, Weinheim 1993, ISBN 3-527-28030-8 , p. 175.
- ↑ PC Nowell, DA Hungerford: A minute chromosome in human granulocytic leukemia. In: Science. 1960; 132, S. 1497 doi: 10.1126 / science.132.3438.1488 Note: The rather short publication is part of a collection of abstracts
- ↑ PC Nowell: The minute chromosome (Phl) in chronic granulocytic leukemia. In: blood. 1962 Apr; 8, pp. 65-66. PMID 14480647
- ↑ PC Nowell: Discovery of the Philadelphia chromosome: a personal perspective. In: J Clin Invest. 2007 Aug; 117 (8), pp. 2033-2035. PMID 17671636 (Free Full-Text)
- ^ JD Rowley: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining . In: Nature. 1973; 243, p. 290. PMID 4126434
- ↑ N. Heisterkamp et al .: Localization of the c-ab1 oncogene adjacent to a translocation break point in chronic myelocytic leukaemia. In: Nature. 1983; 306, pp. 239-242. PMID 6316147
- ↑ J. Groffen et al.: Philadelphia chromosomal breakpoints are clustered within a limited region, bcr, on chromosome 22. In: Cell . 1984; 36, pp. 93-99 PMID 6319012
literature
- Philadelphia chromosome. In: Herder-Lexikon der Biologie. Spektrum Akademischer Verlag, Heidelberg 2003, ISBN 3-8274-0354-5 .
- Bruce Alberts , Dennis Bray, Julian Lewis, Martin Raff , Keith Roberts, James D. Watson: Molecular Biology of the Cell. 1st corrected reprint of the 3rd edition, VCH Verlagsgesellschaft, Weinheim 1997, ISBN 3-527-30055-4 .
- Barbara J. Bain: Leukemia Diagnosis. 2nd Edition. Blackwell Science, Oxford 1999, ISBN 0-632-05165-5 .
- Klaus Czerwenka, Mahmood Manavi, Kerstin Pischinger: Introduction to molecular biology. Verlag Wilhelm Maudrich, Vienna 2003, ISBN 3-85175-796-3 .
- Ricky Lewis: Human Genetics. Concepts and Applications. Wm. C. Brown Publishers, Dubuque 1994, ISBN 0-697-13315-X .
- Orlando J. Miller, Eeva Therman: Human Chromosomes. 4th edition. Springer-Verlag, New York 2001, ISBN 0-387-95046-X .
- Wilhelm Seyffert: Textbook of Genetics. Spectrum Academic Publishing House, 2003.
- Friedrich Vogel, Arno G. Motulsky : Human Genetics. 3. Edition. Springer-Verlag, Berlin / Heidelberg 1997, ISBN 3-540-60290-9 .