DNA binding proteins
A DNA binding protein is a protein that binds to DNA .
properties
DNA-binding proteins are found in all living things and DNA viruses . They have at least one protein domain that can bind to DNA . The bond can be made to various functional groups . The backbone of the DNA consists of alternating phosphate and deoxyribose units. Due to the phosphate groups, the DNA is provided with negative charges in proportion to the chain length , to which protein domains with positively charged amino acids (e.g. lysine , arginine ) or protein domains with negatively charged amino acids with complexed cations (e.g. Mg 2+ or Zn 2+ complexes). Since the deoxyribose phosphate backbone repeats itself continuously, a protein that binds exclusively to the backbone cannot bind to a specific DNA sequence (no sequence specificity). Sequence-specific binding takes place through at least partial binding to a specific sequence of nucleobases ; the backbone can also be partly bound.
Sequence specificity
DNA-binding proteins without sequence specificity are e.g. B. the polymerases, helicases and generally proteins that slide along the DNA (e.g. with DNA clamps). Sequence-specific DNA-binding proteins are e.g. B. transcription factors that trigger gene expression at defined locations ( promoter ) . Due to the higher proportion of nucleobases in the surface, sequence-specific DNA-binding molecules tend to bind in the major groove of the DNA double helix. While some sequence-specific DNA-binding proteins prefer to bind single-stranded DNA (e.g. single-strand-binding protein), others bind double-stranded DNA (most of them) and a few also bind heteroduplexes of DNA and RNA (e.g. telomerase , reverse transcriptases , RNase H ).
Binding of single stranded DNA
Single-stranded DNA occurs permanently on the telomeres in eukaryotes and occurs temporarily in replication , transcription , recombination and DNA repair, e.g. B. the single-strand binding protein and some DNA repair enzymes.
Binding of double stranded DNA
Double-stranded DNA with complementary base pairing forms a double helix ( B-DNA ). This DNA double helix has a large and a small groove. The small groove has a smaller proportion of the nucleobases in the surface of the molecule, which is why it is less suitable for sequence-specific binding. Various DNA-binding molecules such as Lexitropsine , Netropsin , Distamycin , Hoechst 33342 , Pentamidine , DAPI or SYBR Green I bind to the minor groove of double-stranded DNA regardless of the sequence. Double-stranded binding proteins include histones or NS H-DNA-binding protein , high-mobility group , DNA polymerases , DNA-dependent RNA polymerases , helicases , topoisomerases , gyrases , ligases , polynucleotide - kinases , nucleases , some DNA repair enzymes . Sequence-specific dsDNA-binding proteins include transcription factors and some endonucleases . Prokaryotic transcription factors are mostly smaller than the gene expression products they control, while eukaryotic transcription factors are mostly larger than their controlled products and occasionally have multiple copies of a DNA-binding domain.
DNA-binding protein domains
Typical protein domains in dsDNA-binding proteins are the zinc finger domain , the AT hooks , the DNA clamp and the helix-turn-helix motif for DNA binding or the leucine zipper domain (bZIP) for dimerization . In the case of single-stranded DNA, the OB folding domain has been described , among other things .
ID
Methods for determining protein-DNA interactions (bound DNA sequence , DNA-binding proteins) are e.g. B. EMSA , DNase Footprinting Assay , ChIP , DamID , ChIP-on-Chip or ChIP-Seq .
Modeling
Different approaches to molecular modeling have been described. Algorithms for determining the bound DNA sequence have also been developed.
modification
In the course of a protein design , e.g. B. zinc finger proteins or TALENs can be designed. With the CRISPR / Cas method , corresponding DNA sequences can be bound using a complementary RNA sequence.
to form
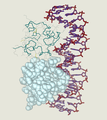
DNA (orange) with histones (blue)
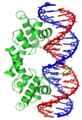
The Lambda - repressor with helix-turn-helix bound to DNA
The restriction enzyme EcoRV (green) with DNA.
Web links
- Abalone tool for modeling DNA-ligand interactions.
- DBD database of predicted transcription factors Uses a curated set of DNA-binding domains to predict transcription factors in all completely sequenced genomes
- MeSH DNA-binding proteins
Individual evidence
- ^ Travers, AA: DNA-protein interactions . Springer, London 1993, ISBN 978-0-412-25990-6 .
- ↑ Pabo CO, Sauer RT: Protein-DNA recognition . In: Annu. Rev. Biochem. . 53, No. 1, 1984, pp. 293-321. doi : 10.1146 / annurev.bi.53.070184.001453 . PMID 6236744 .
- ↑ Dickerson RE: The DNA helix and how it is read . In: Sci Am . 249, No. 6, 1983, pp. 94-111. doi : 10.1038 / scientificamerican1283-94 .
- ↑ Luger K, Mäder A, Richmond R, Sargent D, Richmond T: Crystal structure of the nucleosome core particle at 2.8 A resolution . In: Nature . 389, No. 6648, 1997, pp. 251-60. doi : 10.1038 / 38444 . PMID 9305837 .
- ↑ Li Z, Van Calcar S, Qu C, Cavenee W, Zhang M, Ren B: A global transcriptional regulatory role for c-Myc in Burkitt's lymphoma cells . In: Proc Natl Acad Sci USA . 100, No. 14, 2003, pp. 8164-9. doi : 10.1073 / pnas.1332764100 . PMID 12808131 . PMC 166200 (free full text).
- ↑ Pabo C, Sauer R: Protein-DNA recognition . In: Annu Rev Biochem . 53, No. 1, 1984, pp. 293-321. doi : 10.1146 / annurev.bi.53.070184.001453 . PMID 6236744 .
- ^ TH Dickey, SE Altschuler, DS Wuttke: Single-stranded DNA-binding proteins: multiple domains for multiple functions. In: Structure (London, England: 1993). Volume 21, number 7, July 2013, pp. 1074-1084, doi : 10.1016 / j.str.2013.05.013 . PMID 23823326 . PMC 3816740 (free full text).
- ↑ Iftode C, Y Daniely Borowiec J: Replication Protein A (RPA): the eukaryotic SSB . In: Crit Rev Biochem Mol Biol . 34, No. 3, 1999, pp. 141-80. doi : 10.1080 / 10409239991209255 . PMID 10473346 .
- ↑ J. Kur, M. Olszewski, A. D? Ugo? Ecka, P. Filipkowski: Single-stranded DNA-binding proteins (SSBs) - sources and applications in molecular biology. In: Acta biochimica Polonica. Volume 52, Number 3, 2005, pp. 569-574, PMID 16082412 .
- ↑ Zimmer C, Wähnert U: Nonintercalating DNA-binding ligands: specificity of the interaction and their use as tools in biophysical, biochemical and biological investigations of the genetic material . In: Prog. Biophys. Mol. Biol . 47, No. 1, 1986, pp. 31-112. doi : 10.1016 / 0079-6107 (86) 90005-2 . PMID 2422697 .
- ↑ Dervan PB: Design of sequence-specific DNA-binding molecules . In: Science . 232, No. 4749, April 1986, pp. 464-71. doi : 10.1126 / science.2421408 . PMID 2421408 .
- ↑ Sandman K, Pereira S, Reeve J: Diversity of prokaryotic chromosomal proteins and the origin of the nucleosome . In: Cell Mol Life Sci . 54, No. 12, 1998, pp. 1350-64. doi : 10.1007 / s000180050259 . PMID 9893710 .
- ^ Dame RT: The role of nucleoid-associated proteins in the organization and compaction of bacterial chromatin . In: Mol. Microbiol. . 56, No. 4, 2005, pp. 858-70. doi : 10.1111 / j.1365-2958.2005.04598.x . PMID 15853876 .
- ↑ Thomas J: HMG1 and 2: architectural DNA-binding proteins . In: Biochem Soc Trans . 29, No. Pt 4, 2001, pp. 395-401. doi : 10.1042 / BST0290395 . PMID 11497996 .
- ^ Grosschedl R, Giese K, Pagel J: HMG domain proteins: architectural elements in the assembly of nucleoprotein structures . In: Trends Genet . 10, No. 3, 1994, pp. 94-100. doi : 10.1016 / 0168-9525 (94) 90232-1 . PMID 8178371 .
- ^ Myers L, Kornberg R: Mediator of transcriptional regulation . In: Annu Rev Biochem . 69, No. 1, 2000, pp. 729-49. doi : 10.1146 / annurev.biochem.69.1.729 . PMID 10966474 .
- ^ Spiegelman B, Heinrich R: Biological control throughs regulated transcriptional coactivators . In: Cell . 119, No. 2, 2004, pp. 157-67. doi : 10.1016 / j.cell.2004.09.037 . PMID 15479634 .
- ^ V. Charoensawan, D. Wilson, SA Teichmann: Genomic repertoires of DNA-binding transcription factors across the tree of life. In: Nucleic acids research. Volume 38, Number 21, November 2010, pp. 7364-7377, doi : 10.1093 / nar / gkq617 . PMID 20675356 . PMC 2995046 (free full text).
- ↑ NW Ashton, E. Bolderson, L. Cubeddu, KJ O'Byrne, DJ Richard: Human single-stranded DNA binding proteins are essential for Maintaining genomic stability. In: BMC molecular biology. Volume 14, 2013, p. 9, doi : 10.1186 / 1471-2199-14-9 . PMID 23548139 . PMC 3626794 (free full text).
- ↑ Carey MF, Peterson CL, Smale ST: Experimental strategies for the identification of DNA-binding proteins. In: Cold Spring Harbor protocols. Volume 2012, number 1, January 2012, pp. 18–33, doi : 10.1101 / pdb.top067470 . PMID 22194258 .
- ↑ teif VB, rib K .: Statistical-mechanical lattice models for protein-DNA binding in chromatin. . In: Journal of Physics: Condensed Matter . 2010. arxiv : 1004.5514 .
- ↑ Wong KC, Chan ™, Peng C., Li Y., and Zhang Z. DNA Motif Elucidation using belief propagation , Nucleic Acids Research. Advanced Online June 2013; doi : 10.1093 / nar / gkt574 , PMID 23814189
- ↑ DG Stormo: DNA binding sites: representation and discovery. In: Bioinformatics. Volume 16, Number 1, January 2000, pp. 16-23, PMID 10812473 .
- ↑ Created from PDB 1LMB
- ↑ Created from PDB 1RVA