Reverse transcriptase
| Reverse transcriptase | ||
|---|---|---|
|
||
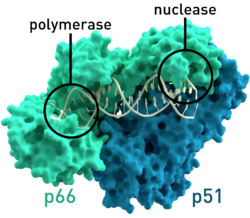
| Reverse transcriptase (dimer) model of HIV- 1 | ||
| Enzyme classification | ||
| EC, category | 2.7.7.49 , nucleotidyl transferase | |
| Substrate | Deoxynucleoside triphosphate + DNA (s) | |
| Products | Diphosphate + DNA (n + 1) | |
Reverse transcriptases ( RT ) are enzymatically active proteins known as RNA-dependent DNA polymerases a transcription , namely in the reverse direction (reverse) of RNA in DNA catalyze ; this allows genetic information to be transcribed from RNA into DNA.
Biochemical aspects
By means of their RNA-dependent DNA polymerase activity, a hybrid double strand of RNA and DNA is first built up after a single-stranded RNA has been presented by linking complementary paired DNA building blocks ( deoxyribonucleotides ). Thereafter, its RNA portion is largely broken down by means of an RNase H activity of a particular section of the protein. The remaining single strand of DNA is finally supplemented to form a double strand of DNA, catalyzed by an additional inherent DNA-dependent DNA polymerase activity of reverse transcriptase.
The error rate of reverse transcriptase due to a lack of proof-reading is 1:10 3 to 1:10 4 and leads to a very high mutation rate .
history
The reverse transcriptase of retroviruses was first in 1970 by both Howard Temin also as independent of David Baltimore described. In 1975 you and Renato Dulbecco received the Nobel Prize in Physiology or Medicine for this discovery . The addition reverse denotes the peculiar ability of this enzyme to build a DNA from an RNA template. The unexpected direction of the process invalidated the previously held doctrine, the so-called central dogma of molecular biology , that the flow of genetic information runs exclusively in the direction of DNA → RNA → protein, never the other way around.
Occurrence
Reverse transcriptase was first discovered in retroviruses (e.g. HIV , HTLV , SIV ). These viruses with an RNA genome use RT to rewrite their genome in DNA. The RT thus fulfills a crucial function in the multiplication of the virus. In addition, certain DNA viruses such as the hepadnaviruses (e.g. the pathogen causing hepatitis B ( HBV, the protein P ) or the caulimoviruses that occur in plants ) also contain an RT. The class I transposons , also known as retro elements , are also derived from former, mutated retroviruses . These need an RT for their replication. This is either coded by you (autonomous LINEs and LTR retrotransposons ) or must be made available (e.g. with SINEs ).
Group II introns also code for a reverse transcriptase, which stabilizes an enzymatically active intron RNA ( ribozyme ) and transcribes the integrated RNA into DNA. The RNA catalyzes the splicing in the process. Group II introns have been detected in prokaryotes and in the genomes of organelles in fungi and plants.
A reverse transcriptase is also part of the telomerase of eukaryotes , where it expands the telomeres, which have been shortened in the course of replication , to their original length and thus delays the process of cell aging . The more precise name is telomerase reverse transcriptase (TERT), such as. B. for human telomerase reverse transcriptase (hTERT).
Reverse transcription
The RNA genome of RNA viruses, for example retroviruses , is transcribed into double-stranded DNA. This process is called reverse transcription . The virus brings the necessary reverse transcriptase with it in its virus particles. With the help of a tRNA primer, this first transcribes the single-stranded RNA of the virus into a complementary DNA strand (activity as an RNA-dependent DNA polymerase ). The RNA is then broken down down to a fragment that serves as a second primer (activity as ribonuclease H ). This then creates the double-stranded DNA (activity as a DNA-dependent DNA polymerase ).
The viral genome of the RNA is then available as a double-stranded DNA copy. In addition, so-called LTR sequences were generated at both ends of the DNA strands during reverse transcription , which are essential for the further course of the infection. They enable integration into the host cell's DNA genome - through another retrovirus enzyme, an integrase .
Biotechnological applications
The frequency of errors in viral reverse transcriptase makes it difficult to combat retroviruses such as HIV. The inhibition of reverse transcriptase is a goal of combination therapy and is possible through various active substances ( NNRTI , NRTI ). Such reverse transcriptase inhibitors were the first and, until 1994, the only effective drugs approved for the treatment of HIV infection.
The artificially generated reverse transcriptase "Xenopolymerase" is now able to proofread and thus minimize the error rate.
Reverse transcriptases are used in molecular biology and in molecular diagnostics , for example in RT-PCR or to create a cDNA bank . For this, viral reverse transcriptases from the Murine Leukemia Virus (MLV) or the Avian Myeloblastosis Virus (AMV) are used. As a rule, however, no native enzymes are used, but genetically engineered variants with a lower error rate, lower RNase H activity and higher temperature stability. After the isolation of recombinant group II intron reverse transcriptases has been successful, it is expected that, due to their lower error rate, higher speed and temperature stability, but in particular also the template switch activity, the direct change to the 3 'end of new RNAs, gain in importance.
Individual evidence
- ^ Racaniello, VR (Vincent R.), Rall, Glenn F., Skalka, Anna Marie, Enquist, LW (Lynn W.) ,: Principles of virology . 4th edition. Washington, DC, ISBN 978-1-55581-933-0 , pp. 189 .
- ↑ a b Reverse Transcriptase with Proofreading Capabilities Created. Retrieved February 27, 2020 (English).
- ↑ HM Temin, S. Mizutani: RNA-dependent DNA polymerase in virions of Rous sarcoma virus. In: Nature. Volume 226, Number 5252, June 1970, pp. 1211-1213, PMID 4316301 .
- ↑ D. Baltimore: RNA-dependent DNA polymerase in virions of RNA tumor viruses. In: Nature. Volume 226, Number 5252, June 1970, pp. 1209-1211, PMID 4316300 .
- ↑ John M. Coffin, Stephen H. Hughes, Harold E. Varmus : The Place of Retroviruses in Biology. In: Retroviruses. Cold Spring Harbor Laboratory Press, 1997, ISBN 0-87969-571-4 .
- ↑ Central dogma reversed. In: Nature. Volume 226, Number 5252, June 1970, pp. 1198-1199, PMID 5422595 .
- ↑ AM Lambowitz, S. Zimmerly: Group II introns: mobile ribozymes that invade DNA. In: Cold Spring Harbor perspectives in biology. Volume 3, number 8, August 2011, p. A003616, doi: 10.1101 / cshperspect.a003616 , PMID 20463000 , PMC 3140690 (free full text) (review).
- ↑ Jeremy Cherfas: Hayflick Licked: Telomerase Lengthens Life of Normal Human Cells ( Memento August 8, 2012 in the Internet Archive )
- ^ Witzany G (August 2008). The Viral Origins of Telomeres and Telomerases and their Important Role in Eukaryogenesis and Genome Maintenance. (PDF; 263 kB). In: Biosemiotics 1 (2): 191-206. doi: 10.1007 / s12304-008-9018-0
- ^ MJ Wacker, MP Godard: Analysis of one-step and two-step real-time RT-PCR using SuperScript III. In: Journal of biomolecular techniques: JBT. Volume 16, Number 3, September 2005, pp. 266-271, PMID 16461951 , PMC 2291734 (free full text).
- ↑ A. Baranauskas, S. Paliksa, G. Alzbutas, M. Vaitkevicius, J. Lubiene, V. Letukiene, p Burinskas, G. Snauvicius, R. Skirgaila: Generation and characterization of new highly thermostable processive table and M-MuLV reverse transcriptase variants. In: Protein engineering, design & selection: PEDS. Volume 25, Number 10, October 2012, pp. 657-668, doi: 10.1093 / protein / gzs034 , PMID 22691702 .
- ^ S. Mohr, E. Ghanem, W. Smith, D. Sheeter, Y. Qin, O. King, D. Polioudakis, VR Iyer, S. Hunicke-Smith, S. Swamy, S. Kuersten, AM Lambowitz: Thermostable group II intron reverse transcriptase fusion proteins and their use in cDNA synthesis and next-generation RNA sequencing. In: RNA. Volume 19, number 7, July 2013, pp. 958-970, doi: 10.1261 / rna.039743.113 , PMID 23697550 , PMC 3683930 (free full text).
Web links
- The reverse transcriptase of HIV
- Interactive presentation of the RT of HIV in the online Macromolecular Museum (the PDB file is displayed via a JavaScript viewer)