Hepatitis B virus
| Hepatitis B virus | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Hepatitis B virus |
||||||||||||||||||
| Systematics | ||||||||||||||||||
|
||||||||||||||||||
| Taxonomic characteristics | ||||||||||||||||||
|
||||||||||||||||||
| Scientific name | ||||||||||||||||||
| Hepatitis B virus | ||||||||||||||||||
| Short name | ||||||||||||||||||
| HBV | ||||||||||||||||||
| Left | ||||||||||||||||||
|
The hepatitis B virus (HBV) is an enveloped DNA virus with a predominantly double-stranded genome and the trigger of the Hepatitis B . The hepatitis B virus comes from the genus Orthohepadnaviruses within the family of the Hepadnaviridae .
illness
In addition to hepatitis B, HBV causes cirrhosis and liver cancer in chronic courses . There may also be a connection in pancreatic cancer . The hepatitis B virus does not have a cytopathic effect , so the damage to the liver cells is increasingly attributed to the immune reaction (indirect pathogenesis , immunopathogenesis ). Through the continuous growth stimulation to replace the infected hepatocytes destroyed by the immune system and through the cytokines , some of which have growth-promoting properties, and through the partial insertion into the cellular DNA, more copying errors occur in the DNA during mitosis (in the S phase), which is what leads to the formation favored by tumors .
construction
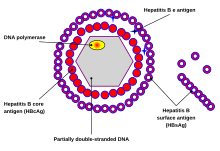
The virion has a virus envelope and an icosahedral capsid . The capsid encloses the viral DNA and a DNA polymerase with a reverse transcriptase activity (RT), similar to a retroviral polymerase. The virus envelope contains HBsAg and the pre-S1 protein as membrane protein for binding to the host cell . With a diameter of 42 nm, the spherical HBV (also called Dane particle ) is one of the smaller animal viruses. There are also non-infectious filamentous and spherical forms without capsid with a diameter of 20 nm, since HBsAg is formed in excess. These virosomes consist of HBsAg in a lipid bilayer . Because of their immunogenicity with no infectivity, they were used in the early hepatitis B vaccines . With recombinant expression (without HBsAg) the HBcAg can also form virosomes.
HBV reaches a virus concentration between 10 4 and 10 9 virions per milliliter in the blood , the virosomes from HBsAg even reach particle concentrations of up to 10 12 particles per milliliter.
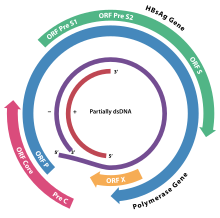
Genome
The HBV genome consists of circularly closed DNA , but the DNA is not completely double-stranded and one strand is continuous. One end of the continuous strand of around 3020–3320 nucleotides binds the viral DNA polymerase, while the shorter strand with 1700 to 2800 nucleotides is significantly shorter.
The continuous (-) strand (non-coding) is reverse complementary to the viral mRNA . The viral DNA can be detected in the cell nucleus shortly after infection . The partially double-stranded DNA is completed on the (+) strand and an RNA sequence on the (+) strand is removed. Finally, overhanging nucleotides are removed and the (-) strand closed to form a ring.
The four genes of HBV are called C ( core ), X, P ( polymerase ) and S ( surface ). The core protein HbcAg is produced from a preprotein by proteolysis . The S gene of the HBsAg protein contains three start codons (AUG) that conform to the reading frame , resulting in three proteins, pre-S1 (synonymous L), pre-S2 (synonymous M) and S.
Various non-coding RNAs have been identified in the HBV genome. These are referred to as Hepatitis B virus PRE alpha , Hepatitis B virus PRE beta and HBV RNA encapsidation signal epsilon .
Proteins
The HBV virion consists of four structural proteins; HBeAg also occurs outside the virion.
HBsAg
HBsAg ( English HBV surface antigen , HBV surface antigen ) is a surface protein of the virion that is recognized as an antigen by antibodies in the course of the immune reaction . It can be detected in the blood of infected people through virological diagnostics and shows when a patient is no longer contagious. In histology, HBsAg is detected by a Shikata orcein stain .
Historically, HBsAg was first known as the Australia Antigen , as it was first discovered in the blood serum of Australian Aborigines by Baruch Samuel Blumberg . Alfred Prince discovered the connection with a form of hepatitis in 1968. The first vaccine against HBV ( Heptavax ) was purified from the blood plasma of those infected with HBV ; due to the risk of contamination by other pathogens , it was later switched to recombinant proteins from baker's yeast .
The hepatitis D virus needs HBsAg to become pathogenic.
HBcAg
HBcAg ( English core antigen , antigen of the virus core ) is used to package the viral DNA and is used to detect a current infection. It is the capsid protein of HBV.
HBeAg
HBeAg ( English extracellular antigen , extracellular antigen) is a splice variant from the open reading frames core-ORF and pre-C-ORF . HBeAg is secreted and is not found in the virion. Like HBcAg, HBeAg is used to detect a current infection. Presumably it serves to reduce the gene expression of toll-like receptor 2 on hepatocytes and monocytes and thus to dampen the immune response against HBV. HBeAg is not essential, ie virions without HBeAg expression are infectious and pathogenic.
Hepatitis B virus DNA polymerase
The DNA polymerase of the HBV is the replication of the viral genome. Lamivudine and penciclovir inhibit polymerase in non-resistant HBV strains.
HBx
Little is known about the function of the X protein. The X protein may be derived from a DNA glycosylase . HBx increases the release of calcium ions from the mitochondria . The HBx changes cellular transcription , the cell cycle and forms a protein complex with the cellular protein HBX interacting protein (HBXIP), which interacts with the spindle apparatus during cell division . HBx also interacts with Damaged DNA Binding Protein 1 (DDB1) and inhibits the CUL4 -DDB1- E3 complexes and thereby the ubiquitin-proteasome system . HBx promotes cell division and thereby promotes the development of tumors. HBx inhibits the protein arginine methyltransferase PRMT1, which increases the replication of HBV.
Occurrence and genotypes
The reservoir host of HBV is mainly the person next to HBV also comes in great apes and gibbons before. The eight genotypes of HBV are named A to H. A genotype I may also exist, but this is not unanimously accepted. The different genotypes respond differently to therapies.
The genotypes differ by at least eight percent at the genome level and have different geographical frequencies. Genotype F differs the most from the others, with a fourteen percent difference. Genotype A is prevalent in Europe , Africa, and South Asia , including the Philippines . Genotypes B and C are predominant in Asia ; the genotype D occurs in the Mediterranean region , the Middle East and India . Genotype E exists in sub- Saharan Africa . The genotype F (or H) occurs in Central and South America . Genotype G was found in France and Germany . Genotypes A, D, and F dominate in Brazil, and all genotypes occur with ethnic clusters in the United States .
Within the genotypes, 24 subtypes are described, which differ from one another in their DNA sequence by four to eight percent.
- Genotype A has two subtypes: Aa (A1) in Africa / Asia and Ae (A2) in Europe / USA.
- Genotype B has two geographically separated subtypes: Bj / B1 ('j' - Japan) and Ba / B2 ('a' - Asia). The genotype Ba is further divided into four clades (B2 - B4).
- Genotype C also has two geographically separated subtypes: Cs (C1) in Southeast Asia and Ce (C2) in East Asia. The C subtype is divided into five clades , while a possible sixth (C6) and a seventh (C7) have so far only been found in one isolate in the Philippines and West Papua. The genotype C1 occurs in Vietnam , Myanmar and Thailand . The genotype C2 occurs in Japan , Korea and China . The C3 genotype is found in New Caledonia and Polynesia . The genotype C4 occurs in Australia and C5 in the Philippines .
- Genotype D is divided into seven subtypes (D1 - D7).
- Genotype E has only one subtype.
- Genotype F is divided into four subtypes (F1 - F4). The subtype F1 is further separated into clades 1a and 1b. In Venezuela , the subtypes F1, F2 and F3 are found in the west and east, while F3 occurs alone in the south. Subtype Ia occurs in Central America, subtype III in northern South America and IV in southern South America. The clade Ib is found in almost the entire American continent (except northern South America ) and the clade II is found in Central and South America.
evolution
The exact determination of the early evolution of HBV is problematic. The splitting of the hepadnaviruses into ortho- and avihepadnaviruses took place approximately 125,000 years ago (95% interval 78,297-313,500). Both avi and ortho hepadnaviruses began to change more rapidly around 25,000 years ago. At this time the division into the eight genotypes took place. Human hepadnaviruses separated approximately 7,000 (95% interval: 5.287–9.270) to 10,000 years (95% interval: 6.305–16.681). The avihepadnaviruses lack the X protein, but residues can still be found in the duck hepadna virus. The rate of non-synonymous HBV mutations was estimated to be about 2 × 10 −5 amino acid substitutions per site per year. The mean frequency of nucleotide substitutions per site per year was estimated at about 7.9 × 10 −5 . Another estimate suspects the last common ancestor of the human HBV strains 1,500 years ago and a separation from the avihepadnaviruses 6,000 years ago with a mutation rate of about 10 −6 substitutions per site per year.
Classification
HBV is a typical representative of the genus Orthohepadnaviruses , which has three other representatives: the Ground squirrel hepatitis virus , the Woodchuck hepatitis virus , and the Woolly monkey hepatitis B virus . The family of the Hepadnaviridae contains two other genera, the Avihepadnaviruses and one to be named. The family is not yet assigned to any order. HBV-like viruses have already been found in all Old World monkeys ( orangutans , gibbons , gorillas and chimpanzees ) and also in New World monkeys such as the woolly monkey , which suggests that this virus originated in primates.
HBV are divided into four serotypes (adr, adw, ayr, ayw), depending on the antigenic epitopes on their coat proteins, genetically they are assigned to eight genotypes (A – H) depending on their mutations . The individual genotypes have a different geographical distribution and are used for the epidemiological determination of transmission and viral evolution. The different genotypes also show differences in the course of the disease , the frequency of complications and in the choice of therapy and vaccine .
Replication cycle
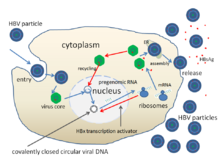
The replication cycle of HBV:
- Low affinity attachment ( adsorption ) to the cell membrane via glycosamine glycans, then binding to NTCP (Na + -taurocholate cotransporting polypeptide) and subsequent endocytosis via a previously unexplained mechanism.
- Penetration (engl. Penetration or uncoating ) by an induced fusion of the viral envelope and the endosomal membrane , whereby the core with the DNA of the virus into the cytosol is liberated.
- Unzip (Engl. Uncoating ) and transport in the cell nucleus via chaperones . This releases the DNA from the core and then completes the (-) strand and closes the ring to form the cccDNA , which is used as a template for the transcription of the four viral mRNAs (Pre-S1 / L, Pre-S2 / M, S and X) and the two RNAs used for expression and genome synthesis (C / P and Pre-C). All viral RNAs have a cap structure and a poly A tail .
- Replication (engl. Replication ) in the cytosol, in which the longest RNA (C / P is longer than the viral DNA genome) serves as a template for the synthesis of the viral DNA genome, as well as protein biosynthesis of the capsid and the viral polymerase. The RNA intermediate increases the mutation rate to generate escape mutations . The cccDNA is imported into the cell nucleus up to the point in time of the synthesis of the envelope proteins HBcAg and HBsAg (after replication); the cccDNA is only exported from the cell nucleus through the envelope proteins. Due to the core import of the genome, the retention of unpackaged viral genomes in the cell nucleus results in the occasional persistent infection .
- Assembly (engl. Assembly ) of the new virions following the pinch-a from the cell or transported back to the cell nucleus where the DNA is separated from the HBcAg and a new genome synthesis begins.
- Release (engl. Budding, release ) of the Tochtervirionen by exocytosis (non- lytic ) with simultaneous synthesis of the DNA genome in the cytoplasm of the longest mRNA by the RT activity of the viral polymerase .
Environmental stability
As an enveloped virus, the hepatitis B virus is sensitive to chemical or physical disinfection processes , but due to the high protein content in the virus envelope, it is only inactivated relatively slowly by alcohols or surfactants compared to other enveloped viruses . The inactivation of HBV by disinfection procedure can with the MAD-test to be determined.
Reporting requirement
In Germany, any direct or indirect evidence of the hepatitis B virus must be reported by name in accordance with Section 7 of the Infection Protection Act .
In Switzerland, the positive laboratory analytical finding is a hepatitis B virus notifiable and that after the Epidemics Act (EpG) in connection with the epidemic Regulation and Annex 3 of the Regulation of EDI on the reporting of observations of communicable diseases of man .
literature
- S. Modrow, Dietrich Falke , U. Truyen: Molecular Virology. 2nd Edition. Spectrum, Heidelberg 2003, ISBN 3-8274-1086-X .
- DM Knipe, Peter M. Howley (Ed.): Fields Virology. 5th edition. Philadelphia 2007, ISBN 978-0-7817-6060-7 .
Individual evidence
- ↑ a b c d e ICTV: ICTV Taxonomy history: Hepatitis B virus , EC 51, Berlin, Germany, July 2019; Email ratification March 2020 (MSL # 35)
- ^ A b M. M. Hassan, D. Li, AS El-Deeb, RA Wolff, ML Bondy, M. Davila, JL Abbruzzese: Association between hepatitis B virus and pancreatic cancer. In: Journal of clinical oncology: official journal of the American Society of Clinical Oncology. Volume 26, Number 28, October 2008, pp. 4557-4562. ISSN 1527-7755 , doi: 10.1200 / JCO.2008.17.3526 , PMID 18824707 , PMC 2562875 (free full text).
- ↑ Richard Hunt: Hepatitis Viruses. ( Memento June 19, 2014 on the Internet Archive ) University of Southern California, Department of Pathology and Microbiology
- ↑ M. Schwalbe, O. Ohlenschläger, A. Marchanka, R. Ramachandran, S. Hafner, T. Heise, M. Görlach: Solution structure of stem-loop alpha of the hepatitis B virus post-transcriptional regulatory element. In: Nucleic acids research. Volume 36, Number 5, March 2008, pp. 1681-1689. ISSN 1362-4962 , doi: 10.1093 / nar / gkn006 , PMID 18263618 , PMC 2275152 (free full text).
- ↑ a b c d e f D. M. Knipe, Peter M. Howley (Ed.): Fields Virology. 5th edition. Philadelphia 2007, ISBN 978-0-7817-6060-7 .
- ↑ FV Chisari, M. Isogawa, SF Wieland: Pathogenesis of hepatitis B virus infection. In: Pathology-Biology. Volume 58, Number 4, August 2010, pp. 258-266, ISSN 1768-3114 . doi: 10.1016 / j.patbio.2009.11.001 . PMID 20116937 . PMC 2888709 (free full text).
- ↑ F. Guerrieri, L. Belloni, N. Pediconi, M. Levrero: Molecular mechanisms of HBV-associated hepatocarcinogenesis. In: Seminars in liver disease. Volume 33, Number 2, May 2013, pp. 147-156, ISSN 1098-8971 . doi: 10.1055 / s-0033-1345721 . PMID 23749671 .
- ↑ S. Locarnini: Molecular virology of hepatitis B virus. In: Seminars in liver disease. Volume 24 Suppl 1, 2004, pp. 3-10. ISSN 0272-8087 , doi: 10.1055 / s-2004-828672 , PMID 15192795 .
- ^ CR Howard: The biology of hepadnaviruses. In: The Journal of general virology. Volume 67 (Pt 7), July 1986, pp. 1215-1235. ISSN 0022-1317 , PMID 3014045 .
- ↑ RS Koff, JT Galambos: Viral hepatitis. In: L. Schiff, ER Schiff (Ed.): Diseases of the liver. 6th edition. JB Lippincott, Philadelphia 1987, pp. 457-581.
- ^ W. Fiers, M. De Filette, K. El Bakkouri, B. Schepens, K. Roose, M. Schotsaert, A. Birkett, X. Saelens: M2e-based universal influenza A vaccine. In: Vaccine. 27 (45), 2009, pp. 6280-6283. PMID 19840661 .
- ↑ A. Kay, F. Zoulim: Hepatitis B virus genetic variability and evolution. In: Virus research. Volume 127, Number 2, August 2007, pp. 164-176 ISSN 0168-1702 , doi: 10.1016 / j.virusres.2007.02.021 , PMID 17383765 .
- ↑ a b J. Beck, M. Nassal: Hepatitis B virus replication. In: World journal of gastroenterology: WJG. Volume 13, Number 1, January 2007, pp. 48-64. ISSN 1007-9327 , PMID 17206754 , PMC 4065876 (free full text).
- ↑ GJ Smith, JE Donello, R. Lueck, G. Steger, TJ Hope: The hepatitis B virus post-transcriptional regulatory element contains two conserved RNA stem-loops Which are required for function. In: Nucleic acids research. Volume 26, Number 21, November 1998, pp. 4818-4827. ISSN 0305-1048 , PMID 9776740 , PMC 147918 (free full text).
- ↑ S. Flodell, J. Schleucher, J. Cromsigt, H. Ippel, K. Kidd-Ljunggren, S. Wijmenga: The apical stem-loop of the hepatitis B virus encapsidation signal folds into a stable tri-loop with two underlying pyrimidines bulges. In: Nucleic acids research. Volume 30, Number 21, November 2002, pp. 4803-4811. ISSN 1362-4962 , PMID 12409471 , PMC 135823 (free full text).
- ↑ P. Guarascio, F. Yentis, U. Cevikbas, B. Portmann, R. Williams: Value of copper-associated protein in diagnostic assessment of liver biopsy. In: Journal of clinical pathology. Volume 36, Number 1, January 1983, pp. 18-23. ISSN 0021-9746 , PMID 6185545 , PMC 498098 (free full text).
- ↑ BS Blumberg, HJ Alter, S. Visnich: A "New" antigen in leukemia sera. In: JAMA. Volume 191, February 1965, pp. 541-546. ISSN 0098-7484 , PMID 14239025 .
- ↑ N. Chai, HE Chang, E. Nicolas, Z. Han, M. Jarnik, J. Taylor: Properties of subviral particles of hepatitis B virus. In: Journal of virology. Volume 82, Number 16, August 2008, pp. 7812-7817. ISSN 1098-5514 , doi: 10.1128 / JVI.00561-08 , PMID 18524834 , PMC 2519590 (free full text).
- ^ Mandell, Douglas, and Bennett's Principles and Practice of Infectious Diseases. 7th edition. Volume 2, New York 2010, p. 2062.
- ↑ SK Ono-Nita, N. Kato, Y. Shiratori, T. Masaki, KH Lan, FJ Carrilho, M. Omata: YMDD motif in hepatitis B virus DNA polymerase influences on replication and lamivudine resistance: A study by in vitro full- length viral DNA transfection. In: Hepatology (Baltimore, Md.). Volume 29, Number 3, March 1999, pp. 939-945. ISSN 0270-9139 , doi: 10.1002 / hep.510290340 . PMID 10051501 .
- ↑ T. Shaw, SS Mok, SA Locarnini: Inhibition of hepatitis B virus DNA polymerase by enantiomers of penciclovir triphosphate and metabolic basis for selective inhibition of HBV replication by penciclovir. In: Hepatology (Baltimore, Md.). Volume 24, Number 5, November 1996, pp. 996-1002. ISSN 0270-9139 , doi: 10.1002 / hep.510240504 , PMID 8903366 .
- ↑ GH Guo, DM Tan, PA Zhu, F. Liu: Hepatitis B virus X protein promotes proliferation and upregulates TGF-beta1 and CTGF in human hepatic stellate cell line, LX-2. In: Hepatobiliary & pancreatic diseases international: HBPD INT. Volume 8, Number 1, February 2009, pp. 59-64. ISSN 1499-3872 , PMID 19208517 .
- ↑ MJ Bouchard, RJ Schneider: The enigmatic X gene of hepatitis B virus. In: Journal of virology. Volume 78, number 23, December 2004, pp. 12725-12734 ISSN 0022-538X , doi: 10.1128 / JVI.78.23.12725-12734.2004 , PMID 15542625 , PMC 524990 (free full text).
- ↑ a b c F. J. van Hemert, MA van de Klundert, VV Lukashov, NA Kootstra, B. Berkhout, HL Zaaijer: Protein X of hepatitis B virus: origin and structure similarity with the central domain of DNA glycosylase. In: PloS one. Volume 6, number 8, 2011, p. E23392 ISSN 1932-6203 , doi: 10.1371 / journal.pone.0023392 . PMID 21850270 , PMC 3153941 (free full text).
- ↑ SL McClain, AJ Clippinger, R. Lizzano, MJ Bouchard: Hepatitis B virus replication is associated with an HBx-dependent mitochondrion-regulated increase in cytosolic calcium levels. In: Journal of virology. Volume 81, Number 21, November 2007, pp. 12061-12065. ISSN 0022-538X , doi: 10.1128 / JVI.00740-07 . PMID 17699583 , PMC 2168786 (free full text).
- ↑ MJ Bouchard, RJ Puro, L. Wang, RJ Schneider: Activation and inhibition of cellular calcium and tyrosine kinase signaling pathways identify targets of the HBx protein involved in hepatitis B virus replication. In: Journal of virology. Volume 77, Number 14, July 2003, pp. 7713-7719. ISSN 0022-538X , PMID 12829810 , PMC 161925 (free full text).
- ↑ Y. Wen, VS Golubkov, AY Strongin, W. Jiang, JC Reed: Interaction of hepatitis B viral oncoprotein with cellular target HBXIP dysregulates centrosome dynamics and mitotic spindle formation. In: The Journal of biological chemistry. Volume 283, Number 5, February 2008, pp. 2793-2803. ISSN 0021-9258 , doi: 10.1074 / jbc.M708419200 . PMID 18032378 .
- ↑ T. Li, EI Robert, PC van Breugel, M. Strubin, N. Zheng: A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery. In: Nature structural & molecular biology. Volume 17, Number 1, January 2010, pp. 105-111. ISSN 1545-9985 , doi: 10.1038 / nsmb.1719 , PMID 19966799 , PMC 2823288 (free full text).
- ↑ MC Kew: Hepatitis B virus x protein in the pathogenesis of hepatitis B virus-induced hepatocellular carcinoma. In: Journal of Gastroenterology and Hepatology . Volume 26 Suppl 1, January 2011, pp. 144-152. ISSN 1440-1746 , doi: 10.1111 / j.1440-1746.2010.06546.x , PMID 21199526 .
- ↑ S. Benhenda, A. Ducroux, L. Rivière, B. Sobhian, MD Ward, S. Dion, O. Hantz, U. Protzer, ML Michel, M. Benkirane, OJ Semmes, MA Buendia, C. Neuveut: Methyltransferase PRMT1 is a binding partner of HBx and a negative regulator of hepatitis B virus transcription. In: Journal of virology. Volume 87, Number 8, April 2013, pp. 4360-4371. ISSN 1098-5514 , doi: 10.1128 / JVI.02574-12 , PMID 23388725 , PMC 3624337 (free full text).
- ↑ P. Sa-Nguanmoo, P. Rianthavorn, S. Amornsawadwattana, Y. Poovorawan: Hepatitis B virus infection in non-human primates. In: Acta virologica. Volume 53, Number 2, 2009, pp. 73-82, ISSN 0001-723X . PMID 19537907 .
- ↑ a b A. Kramvis, M. Kew, G. François: Hepatitis B virus genotypes. In: Vaccine. Volume 23, Number 19, March 2005, pp. 2409-2423. ISSN 0264-410X , doi: 10.1016 / j.vaccine.2004.10.045 , PMID 15752827 .
- ↑ CM Olinger, P. Jutavijittum, JM Hübschen, A. Yousukh, B. Samountry, T. Thammavong, K. Toriyama, CP Muller: Possible new hepatitis B virus genotype, southeast Asia. In: Emerging infectious diseases. Volume 14, number 11, November 2008, pp. 1777-1780 ISSN 1080-6059 , doi: 10.3201 / eid1411.080437 , PMID 18976569 , PMC 2630741 (free full text).
- ↑ F. Kurbanov, Y. Tanaka, A. Kramvis, P. Simmonds, M. Mizokami: When should "I" consider a new hepatitis B virus genotype? In: Journal of virology. Volume 82, Number 16, August 2008, pp. 8241-8242. ISSN 1098-5514 , doi: 10.1128 / JVI.00793-08 . PMID 18663008 , PMC 2519592 (free full text).
- ^ MA Mahtab, S. Rahman, M. Khan, F. Karim: Hepatitis B virus genotypes: an overview. In: Hepatobiliary & pancreatic diseases international: HBPD INT. Volume 7, Number 5, October 2008, pp. 457-464. ISSN 1499-3872 , PMID 18842489 .
- ↑ L. Cavinta, J. Sun, A. May, J. Yin, M. von Meltzer, M. Radtke, NG Barzaga, G. Cao, S. Schaefer: A new isolate of hepatitis B virus from the Philippines possibly representing a new subgenotype C6. In: Journal of medical virology. Volume 81, Number 6, June 2009, pp. 983-987. ISSN 1096-9071 , doi: 10.1002 / jmv.21475 . PMID 19382274 .
- ↑ MI Lusida, VE Nugrahaputra, Soetjipto, R. Handajani, M. Nagano-Fujii, M. Sasayama, T. Utsumi, H. Hotta: Novel subgenotypes of hepatitis B virus genotypes C and D in Papua, Indonesia. In: Journal of clinical microbiology. Volume 46, Number 7, July 2008, pp. 2160-2166. ISSN 1098-660X , doi: 10.1128 / JCM.01681-07 , PMID 18463220 , PMC 2446895 (free full text).
- ^ B. Lin, DA Anderson: A vestigial X open reading frame in duck hepatitis B virus. In: Intervirology. Volume 43, number 3, 2000, pp. 185-190 ISSN 0300-5526 , doi: 10.1159 / 000025037 , PMID 11044813 .
- ↑ C. Osiowy, E. Giles, Y. Tanaka, M. Mizokami, GY Minuk: Molecular evolution of hepatitis B virus over 25 years. In: Journal of virology. Volume 80, Number 21, November 2006, pp. 10307-10314. ISSN 0022-538X , doi: 10.1128 / JVI.00996-06 , PMID 17041211 , PMC 1641782 (free full text).
- ^ Y. Zhou, EC Holmes: Bayesian estimates of the evolutionary rate and age of hepatitis B virus. In: J Mol Evol. 65 (2), 2007, pp. 197-205.
- ↑ Mason, WS, et al .: 00.030. Hepadnaviridae. In: ICTVdB Index of Viruses. International Committee on Taxonomy of Viruses, July 8, 2008, accessed March 13, 2009 .
- ↑ LO Magnius, H. Norder: Subtypes, genotypes and molecular epidemiology of the hepatitis B virus as reflected by sequence variability of the S-gene. In: Intervirology. Volume 38, Numbers 1-2, 1995, pp. 24-34. ISSN 0300-5526 , PMID 8666521
- ↑ M. Levrero, T. Pollicino, J. Petersen, L. Belloni, G. Raimondo, M. Dandri: Control of cccDNA function in hepatitis B virus infection. In: Journal of hepatology. Volume 51, Number 3, September 2009, pp. 581-592, ISSN 1600-0641 , doi: 10.1016 / j.jhep.2009.05.022 , PMID 19616338 .
- ^ V. Bruss: Hepatitis B virus morphogenesis. In: World journal of gastroenterology: WJG. Volume 13, Number 1, January 2007, pp. 65-73. ISSN 1007-9327 , PMID 17206755 , PMC 4065877 (free full text).
- ↑ O. Thraenhart, EK Kuwert, N. Scheiermann, R. Dermietzel, D. Paar, D. Maruhn, A. Alberti, HJ Richter, J. Hotz: Comparison of the morphological alteration and disintegration test (MADT) and the chimpanzee infectivity test for determination of hepatitis B virucidal activity of chemical disinfectants. In: Central Journal for Bacteriology, Microbiology and Hygiene. 1. Dept. Originals B, Hygiene. Volume 176, Numbers 5-6, 1982, pp. 472-484, ISSN 0174-3015 , PMID 7158127 .