BamHI
| BamHI | ||
|---|---|---|
|
||
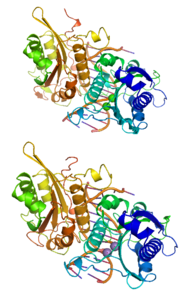
| Ribbon model of a BamHI dimer combined with DNA before (above) and after (below) the cleavage of a DNA strand. The cleavage site is located in the immediate vicinity of the calcium (green, above) and manganese (purple, below) cations. | ||
| Mass / length primary structure | 213 amino acids | |
| Secondary to quaternary structure | Homodimer | |
| Cofactor | 2 mg 2+ | |
| Identifier | ||
| Gene name (s) | R.BamHI | |
| External IDs | ||
| Enzyme classification | ||
| EC, category | 3.1.21.4 , restriction enzyme | |
| Response type | hydrolysis | |
| Substrate | DNA | |
| Products | Double stranded DNA fragments with terminal 5 'phosphate groups | |
BamHI (also BamI ) is an enzyme that is used in molecular biology for the targeted cleavage of DNA . This enzyme belongs to the family of type II restriction enzymes and was first obtained in 1975 from the bacterium Bacillus amyloliquefaciens . BamHI has a molar mass of about 25 kDa . The presence of magnesium ions is essential for enzymatic activity. After dimerization , the enzyme cuts double-stranded DNA within the palindromic recognition sequence to form a 5 'overhang as follows:
| Recognition sequence | Restriction cut |
|---|---|
5'-GGATCC-3' 3'-CCTAGG-5' |
5'-G GATCC-3' 3'-CCTAG G-5' |
This formation of a 5 'overhang ( sticky ends ) comprising four nucleobases by BamHI is used in molecular biology to facilitate the linking ( ligation ) of DNA fragments. BamHI is relatively heat-stable and, under certain conditions, shows a reduced selectivity for the recognition sequence ( star activity ).
Individual evidence
- ↑ Wilson GA & Young FE (1975): Isolation of a sequence-specific endonuclease (BamI) from Bacillus amyloliquefaciens H. In: J. Mol. Biol. 97: 123-125.
- ↑ George J. & Chirikjian JG (1982): Sequence-specific endonuclease BamHI: Relaxation of sequence recognition. In: Proc. Natl. Acad. Sci. USA 79: 2432-2436.