Directed evolution
As directed evolution ( English directed evolution ) refers to the optimization and modification of proteins , enzymes and nucleic acids by mimicking the natural selection . By directed evolution can by means of a random-based mutagenesis and subsequent selection , mutants are identified with improved properties. It is mainly used in white biotechnology and biocatalysis .
principle
Random-based mutagenesis is mostly carried out using PCR- based methods such as error-prone PCR . There are also recombinative methods such as DNA shuffling , Structure-Based Combinatorial Protein Engineering (SCOPE) or RACHITT , in which a chimeric gene is generated from two or more related genes.
The identification of improved mutants of the protein is crucial for the success of directed evolution. The analysis of successful mutants often produces surprising results, such as mutations which have a decisive influence on the activity of the enzyme, even though they are very far outside the active center of the enzyme. Directed evolution contributes so much to understanding how enzymes work.
Frances H. Arnold , who received the Nobel Prize in Chemistry in 2018 for her work , coined the first law of random mutation and selection: "You get what you screen for" (you get (only) what you are looking for). The right choice of fitness function is crucial for the success of directed evolution.
While rational protein design , i.e. changing a protein through the targeted introduction of mutations into its gene , requires precise knowledge of the structure of the protein and its mechanism of action (in the case of enzymes, the reaction mechanism ), directed evolution only requires selection of the mutants sought . Directed evolution is therefore the method of choice for properties of enzymes such as solvent or temperature stability, the molecular causes of which are not yet sufficiently understood. Further applications are changing the regioselectivity , chemoselectivity and enantioselectivity of enzymes as well as the substrate specificity , in particular towards unnatural substrates . The directed evolution of nucleic acids is mainly used in aptamer technology ( SELEX ) and for the optimization of ribozymes .
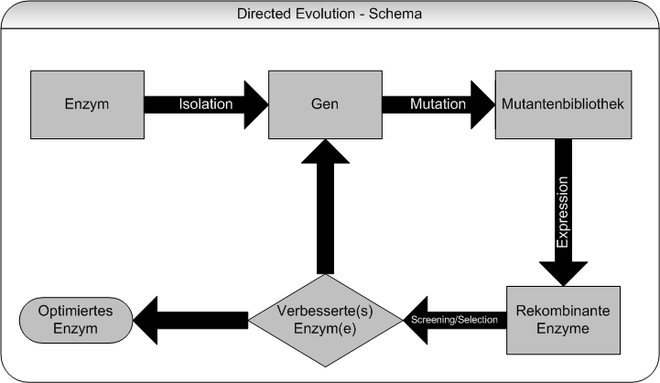
flow chart
application
The Tre recombinase was generated by directed evolution.
literature
- MT Reetz, JD Carballeira: Iterative saturation mutagenesis (ISM) for rapid directed evolution of functional enzymes . In: Nat Protoc. . 2, No. 4, 2007, pp. 891-903. doi : 10.1038 / nprot.2007.72 .
- Rolf D. Schmid: Pocket Atlas of Biotechnology and Genetic Engineering. Weinheim: Wiley-VCH, 2002, ISBN 3-527-30865-2
- Susanne Brakmann, Kai Johnsson: Directed molecular evolution of proteins or how to improve enzymes for biocatalysis. Wiley-VCH, Weinheim 2002, ISBN 3-527-30423-1
- L. Yuan, I. Kurek et al. a .: Laboratory-directed protein evolution. In: Microbiology and molecular biology reviews: MMBR. Volume 69, Number 3, September 2005, pp. 373-392, ISSN 1092-2172 . doi: 10.1128 / MMBR.69.3.373-392.2005 . PMID 16148303 . PMC 1197809 (free full text). (Review).
- Xiaoman Li, Ziding Zhang, Jiangning Song: Computational protein design approaches with significant biological outcomes: progress and challenges. In: Computational and Structural Biotechnology Journal. 2, 2012, doi: 10.5936 / csbj.201209007 . (Review)
Web links
- the Hilvert Lab: " Directed Evolution ", ETH Zurich
Individual evidence
- ^ Stefan Lutz: Beyond directed evolution — semi-rational protein engineering and design . In: Current Opinion in Biotechnology . tape 21 , no. 6 , December 2010, p. 734–743 , doi : 10.1016 / j.copbio.2010.08.011 , PMID 20869867 (English, elsevier.com [accessed June 5, 2019]).
- ^ Frances H. Arnold: Design by directed evolution. Acc. Chem. Res. (1998) Vol. 31, pp. 125-131.