Protein fragment complementation assay
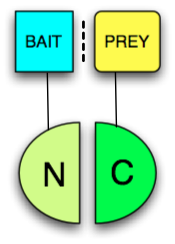
A protein-fragment complementation assay , PCA for short , is a biochemical method for the detection of in vivo protein-protein interactions .
principle
A PCA is a method of detecting binding by complementing fragments of a reporter protein that has previously been broken down into two inactive parts. One part is attached to each of the two proteins to be examined as a fusion protein , so that when the two proteins bind, the attached parts are reconstituted and the reporter protein becomes active. One fragment is coupled as a fusion protein to a bait protein , the second fragment to a prey protein .
- Bimolecular fluorescence complementation
- Dihydrofolate reductase (DHFR)
- Beta-lactamase
- Yeast Gal4 (as in the Yeast Two-Hybrid System )
- Split TEV ( Tobacco etch virus - Protease )
- Luciferase
- Ubiquitin
- Green fluorescent protein (split GFP), e.g. B. EGFP
- LacZ ( beta-galactosidase )
application
The enzyme dihydrofolate reductase, which is divided into two protein fragments that are individually non-functional, is used for the in vivo selection of antibodies . The gene segment that codes for part of the enzyme is fused with the genetic information of an antigen . As a result, this part of the enzyme receives the antigen as a protein tag . The other part of the enzyme now receives an antibody fragment in a similar way, attached in the form of a library . If one of the random antibodies now binds to the antigen, the two halves of the enzyme are brought back together close enough that they can form an active enzyme. Since dihydrofolate reductase is an enzyme that bacterial cells need to survive, only those cells will survive that have a suitable antibody to the antigen. These cells are then usually multiplied and the gene segment that codes for the antibody is sequenced in order to examine the antibody more closely.
Individual evidence
- ↑ K. Tarassov, V. Messier, CR Landry, S. Radinovic, MM Serna Molina, I. Shames, Y. Malitskaya, J. Vogel, H. Bussey, SW Michnick: An in vivo map of the yeast protein interactome. In: Science. Volume 320, number 5882, June 2008, pp. 1465-1470, doi : 10.1126 / science.1153878 . PMID 18467557 .
- ↑ JH Park, JH Back, SH Hahm, HY Shim, MJ Park, SI Ko, YS Han: Bacterial beta-lactamase fragmentation complementation strategy can be used as a method for identifying interacting protein pairs. In: Journal of microbiology and biotechnology. Volume 17, Number 10, October 2007, pp. 1607-1615, PMID 18156775 .
- ^ I. Remy, G. Ghaddar, SW Michnick: Using the beta-lactamase protein-fragment complementation assay to probe dynamic protein-protein interactions. In: Nature protocols. Volume 2, number 9, 2007, pp. 2302-2306, doi : 10.1038 / nprot.2007.356 . PMID 17853887 .
- ↑ MC Wehr, R. Laage, U. Bolz, TM Fischer, S. Grünewald, S. Scheek, A. Bach, KA Nave, MJ Rossner: Monitoring regulated protein-protein interactions using split TEV. In: Nature methods. Volume 3, Number 12, December 2006, pp. 985-993, doi : 10.1038 / nmeth967 . PMID 17072307 .
- ↑ P. Cassonnet, C. Rolloy, G. Neveu, PO Vidalain, T. Chantier, J. Pellet, L. Jones, M. Muller, C. Demeret, G. Gaud, F. Vuillier, V. Lotteau, F. Tangy, M. Favre, Y. Jacob: Benchmarking a luciferase complementation assay for detecting protein complexes. In: Nature methods. Volume 8, Number 12, December 2011, pp. 990-992, doi : 10.1038 / nmeth.1773 . PMID 22127214 .
- ↑ A. Dünkler, J. Müller, N. Johnsson: Detecting protein-protein interactions with the split-ubiquitin sensor. In: Methods in molecular biology (Clifton, NJ). Volume 786, 2012, pp. 115-130, doi : 10.1007 / 978-1-61779-292-2_7 . PMID 21938623 .
- ↑ E. Barnard, DJ Timson: Split-EGFP screens for the detection and localization of protein-protein interactions in living yeast cells. In: Methods in molecular biology (Clifton, NJ). Volume 638, 2010, pp. 303-317, doi : 10.1007 / 978-1-60761-611-5_23 . PMID 20238279 .
- ↑ BD Blakeley, AM Chapman, BR McNaughton: Split-superpositive GFP reassembly is a fast, efficient, and robust method for detecting protein-protein interactions in vivo. In: Molecular bioSystems. Volume 8, Number 8, August 2012, pp. 2036-2040, doi : 10.1039 / c2mb25130b . PMID 22692102 .
- ↑ S. Cabantous, HB Nguyen, JD Pedelacq, F. Koraïchi, A. Chaudhary, K. Ganguly, MA Lockard, G. Favre, TC Terwilliger, GS Waldo: A new protein-protein interaction sensor based on tripartite split-GFP association . In: Scientific reports. Volume 3, 2013, p. 2854, doi : 10.1038 / srep02854 . PMID 24092409 . PMC 3790201 (free full text).
- ^ F. Rossi, CA Charlton, HM Blau: Monitoring protein-protein interactions in intact eukaryotic cells by beta-galactosidase complementation. In: Proceedings of the National Academy of Sciences . Volume 94, Number 16, August 1997, pp. 8405-8410, PMID 9237989 . PMC 22934 (free full text).
- ^ H. Koch, N. Gräfe, R. Schiess & A. Plückthun (2006): Direct Selection of Antibodies from Complex Libraries with the Protein Fragment Complementation Assay . In: J. Mol. Biol. Vol. 357, pp. 427-441. PMID 16442560 doi : 10.1016 / j.jmb.2005.12.043