Assemblin
| Dimer assemblin | ||
|---|---|---|

|
||
| Dimeric pseudorabies virus assemblin (ΔAla225), inhibited with DFP ( space - filling model ). Dashed line: dimer axis, violet: catalytic triad, cyan: oxyanion-hole loop , according to PDB 4V08 | ||
|
Existing structure data : 4V07 , 4V08 , 1CMV , 1VZV , 1AT3 , 1O6E , 1FL1 , 2PBK |
||
| Mass / length primary structure | 24.5 kDa or 225 amino acids per monomer ( pseudorabies virus ) | |
| Secondary to quaternary structure | Homodimer | |
| Identifier | ||
| Gene name (s) | UL26 | |
| External IDs | ||
| Enzyme classification | ||
| EC, category | 3.4.21.97 , serine protease | |
| MEROPS | S21.001 | |
| Response type | hydrolysis | |
| Substrate | a) assembly protein b) scaffold protein | |
| Products | a) assemblin, scaffold protein with linker, mature scaffold protein with linker, main capsid protein binding site b) mature scaffold protein, main capsid protein binding site | |
| Occurrence | ||
| Parent taxon | Herpes viral | |
| Orthologue | ||
| HSV-1 | HCMV | |
| Entrez | 2703453 | 3077485 |
| UniProt | P10210 | P16753 |
| Refseq (protein) | YP_009137100.1 | YP_081529.1 |
| PubMed search | 2703453 |
3077485
|
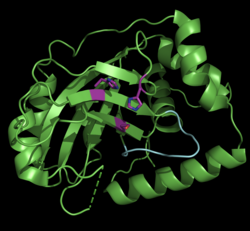
| Monomeric assemblin | ||
|---|---|---|
|
||
| Monomeric pseudorabies virus assemblin (ΔAla225), violet: catalytic triad, cyan: oxyanion-hole loop , according to PDB 4V0T (chain A) | ||
| Mass / length primary structure | 24.5 kDa , 225 amino acids ( pseudorabies virus ) | |
| Secondary to quaternary structure | Monomer | |
Assemblins are a class of serine proteases that are found in all herpes viruses and have a unique catalytic triad . This consists of a serine and two histidines. The activity of the assembline is essential for virus replication and is regulated by a monomer - dimer balance. The weakly associated dimer ( K D in the micromolar range) is the active species. The equilibrium position depends on the assemblin and salt concentration.
nomenclature
The name "assemblin" is derived from its function of cutting the assembly protein (assemblin connected to the scaffold protein via a linker). The nomenclature of the assembline, scaffold proteins and the assembly protein is inconsistent in the literature, as these proteins were and are named according to different criteria. For example, the HCMV assemblin was initially referred to as "VP24" (viral protein, 24 kDa).
Further general names for the protease are "Pr", " maturational protease ", " capsid protease " or simply " protease ". The latter name can be misleading, as there is at least one other protease in herpes viruses. Occasionally the scaffold protein or the assembly protein is incorrectly referred to as assemblin.
function
During the capsid assembly , scaffold proteins accumulate autocatalytically and form a fragile, spherical procapsid with a defined diameter. The protein structure consists of two gene products, the assembly protein and the structure protein, which are present in a ratio of 1:10. The assembly protein consists of two domains : an N-terminal serine protease domain (assemblin) and a C-terminal scaffold protein domain, which are linked by a linker region . The scaffold protein corresponds to the N-terminal domain of the assembly protein . In the procapsid, the assemblind domains dimerize due to the spatial proximity to one another. This activates the assemblin and processes the assembly protein and the scaffold protein. The capsid matures, ie structural changes transform the fragile, spherical procapsid into the more stable icosahedral capsid. The viral DNA is then pumped through the portal into the mature capsids with the help of the terminase complex and cut to the length of the genome. In the course of this, the scaffold protein is displaced from the capsid, while the assemblin remains in the nucleocapsid.
Interestingly, the activity of free assemblin seems to be lower than that of assemblin as part of the assembly protein .
structure
The active, dimeric structure of various (especially human) herpes viruses is known. The monomeric structure of the pseudorabies virus assembly, shortened by one amino acid at the C-terminal, is also known.
The protein consists of a β-barrel structure, which is formed by two β-sheets . The β-barrel is surrounded by 6–9 α-helices . Each monomer carries a complete catalytic triad that is solvent-accessible. The dimer interface is formed by 3 α-helices per monomer and is approximately 1,300 A². The order of the dimerization area is significantly increased by the dimerization (disorder-to-order mechanism). This leads to a shift of a loop ( oxyanion-hole loop ), which in the dimer conformation forms the oxyanion hole with its peptide backbone . The position and orientation of the catalytic triad remains almost unchanged.
In monomeric assemblies of beta and gamma herpes viruses, the two C-terminal helices are disordered, while this is not the case with alpha herpes viruses.
Interfaces
All assemblines cut in at least two different places. They cut autoproteolytically at the so-called R and M sites ( release or maturational ) of the assembly protein and proteolytically at the M site of the scaffold protein. The R-site is located between the assembly protein and the framework protein domain of the assembly protein . The M-site is at the N-terminus of the scaffold protein domain, where the major capsid protein is bound to the scaffold protein.
The consensus sequence of the R-site is Y - V / L - K / Q - A | S / N / T, where P1 'is mostly serine. The consensus sequence of the M-site is V / L / I - X - A | S. X is Asn, Gln, Asp or Glu. Investigations on the HCMV assemblin showed that the M-site is cut faster than the R-site .
In the assembly proteins of some herpes viruses (e.g. HCMV) there are other interfaces for the assemblin, which are presumably used for regulation.
Inhibitors
Most classic serine protease inhibitors hardly inhibit assemblins. Only DFP inhibits assembline. This happens through a covalent bond to the serine of the active center. In addition, helical peptide mimetics have been found which bind non-covalently to the dimerization surface of the monomer and prevent dimerization and thus activation of the protein.
Individual evidence
- ^ Waxman L, Darke PL: The herpesvirus proteases as targets for antiviral chemotherapy . In: Antivir. Chem. Chemother. . 11, No. 1, January 2000, pp. 1-22. doi : 10.1177 / 095632020001100101 . PMID 10693650 .
- ↑ Darke PL, Cole JL, Waxman L, Hall DL, Sardana MK, Kuo LC: Active human cytomegalovirus protease is a dimer . In: J. Biol. Chem. . 271, No. 13, March 1996, pp. 7445-7449. doi : 10.1074 / jbc.271.13.7445 . PMID 8631772 .
- ↑ a b c Zühlsdorf M, values S, Klupp BG, Palm GJ, Mettenleiter TC, Hinrichs W: Dimerization-Induced Allosteric Changes of the Oxyanion-Hole Loop Activate the Pseudorabies Virus Assemblin pUL26N, a Herpesvirus Serine Protease . In: PLoS Pathog. . 11, No. 7, July 2015. doi : 10.1371 / journal.ppat.1005045 . PMID 26161660 . PMC PMC4498786 (free full text).
- ↑ Wang S, Wang K, Li J, Zheng C: Herpes simplex virus 1 ubiquitin-specific protease UL36 inhibits beta interferon production by deubiquitinating TRAF3. . In: J. Virol. . 87, No. 21, August 2013, pp. 11851-11860. doi : 10.1128 / JVI.01211-13 . PMID 23986588 . PMC PMC3807349 (free full text).
- ↑ Newcomb WW, Homa FL, Thomsen DR, Trus BL, Cheng N, Steven A, Booy F, Brown JC: Assembly of the herpes simplex virus procapsid from purified components and identification of small complexes containing the major capsid and scaffolding proteins . In: J. Virol. . 73, No. 5, May 1999, pp. 4239-4250. PMID 10196320 . PMC PMC104203 (free full text).
- ↑ Sheaffer AK, Newcomb WW, Brown JC, Gao M, Weller SK, Tenney DJ: Evidence for controlled incorporation of herpes simplex virus type 1 UL26 protease into capsids . In: J. Virol. . 74, No. 15, August 2000, pp. 6838-6848. doi : 10.1128 / JVI.74.15.6838-6848.2000 . PMID 10888623 . PMC PMC112201 (free full text).
- ^ Newcomb WW, Trus BL, Booy FP, Steven AC, Wall JS, Brown JC: Structure of the herpes simplex virus capsid. Molecular composition of the pentons and the triplexes. . In: J. Mol. Biol. . 232, No. 2, July 1993, pp. 499-511. doi : 10.1006 / jmbi.1993.1406 . PMID 8393939 .
- ↑ Nomura AM, Marnett AB, Shimba N, Dötsch V, Craik CS: Induced structure of a helical switch as a mechanism to regulate enzymatic activity . In: Nat. Struct. Mol. Biol . 12, No. 11, November 2005, pp. 1019-1020. doi : 10.1038 / nsmb1006 . PMID 16244665 .
- Jump up ↑ Robertson BJ, McCann PJ 3rd, Matusick-Kumar L, Newcomb WW, Brown JC, Colonno RJ, Gao M: Separate functional domains of the herpes simplex virus type 1 protease: evidence for cleavage inside capsids . In: Nat. Struct. Mol. Biol . 70, No. 7, July 1996, pp. 4317-4328. PMID 8676454 . PMC PMC190364 (free full text).
- ^ Trus BL, Booy FP, Newcomb WW, Brown JC, Homa FL, Thomsen DR, Steven AC: The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly . In: J. Mol. Biol. . 263, No. 3, November 1996, pp. 447-462. doi : 10.1016 / S0022-2836 (96) 80018-0 . PMID 8918600 .
- ^ Heymann JB, Cheng N, Newcomb WW, Trus BL, Brown JC, Steven AC: Dynamics of herpes simplex virus capsid maturation visualized by time-lapse cryo-electron microscopy . In: Nat. Struct. Biol . 10, No. 5, May 2003, pp. 334-341. doi : 10.1038 / nsb922 . PMID 12704429 .
- ↑ Beard PM, Taus NS, Baines JD: DNA Cleavage and Packaging Proteins Encoded by Genes UL28, UL15, and UL33 of Herpes Simplex Virus Type 1 Form a Complex in Infected Cells . In: J. Virol. . 76, No. 10, May 2002, pp. 4785-4791. doi : 10.1128 / JVI.76.10.4785-4791.2002 . PMID 11967295 . PMC PMC136146 (free full text).
- ↑ White CA, Stow ND, Patel AH, Hughes M, Preston VG: Herpes Simplex Virus Type 1 Portal Protein UL6 Interacts with the Putative Terminase Subunits UL15 and UL28 . In: J. Virol. . 77, No. 11, June 2003, pp. 6351-6358. doi : 10.1128 / JVI.77.11.6351-6358.2003 . PMID 12743292 . PMC PMC154995 (free full text).
- ↑ a b Fernandes SM, Brignole EJ, Taori K, Gibson W: Cytomegalovirus Capsid Protease: Biological Substrates Are Cleaved More Efficiently by Full-Length Enzyme (pUL80a) than by the Catalytic Domain (Assemblin) . In: J. Virol. . 85, No. 7, April 2011, pp. 3526-3534. doi : 10.1128 / JVI.02663-10 . PMID 21270147 . PMC PMC3067851 (free full text).
- ↑ Lee GM, Shahian T, Baharuddin A, Gable JE, Craik CS: Enzyme inhibition by allosteric capture of an inactive conformation . In: J. Mol. Biol. . 411, No. 5, September 2011, pp. 999-1016. doi : 10.1016 / j.jmb.2011.06.032 . PMID 21723875 . PMC PMC3157250 (free full text).
- ↑ Nomura AM, Marnett AB, Shimba N, Dötsch V, Craik CS: Induced structure of a helical switch as a mechanism to regulate enzymatic activity . In: Nat. Struct. Mol. Biol . 12, No. 11, November 2005, pp. 1019-1020. doi : 10.1038 / nsmb1006 . PMID 16244665 .
- ↑ Preston VG, Rixon FJ, McDougall IM, McGregor M, al Kobaisi MF: Processing of the herpes simplex virus protein ICP35 assembly near its carboxy terminal end requires the product of the whole of the UL26 reading frame . In: Virol. . 186, No. 1, January 1992, pp. 87-98. doi : 10.1016 / 0042-6822 (92) 90063-U . PMID 1309284 .
- ↑ Stevens JT, Mapelli C, Tsao J, Hail M, O'Boyle D 2nd, Weinheimer SP, Diianni CL: In vitro proteolytic activity and active-site identification of the human cytomegalovirus protease . In: Eur. J. Biochem. . 226, No. 2, December 1994, pp. 361-367. doi : 10.1111 / j.1432-1033.1994.tb20060.x . PMID 8001553 .
- ↑ Gibson W, Welch AR, Hall MRT: Assemblin, a herpes virus serine maturational proteinase and new molecular target for antivirals . In: Perspect. Drug Discov. Of. . 2, No. 3, July 1995, pp. 413-426. doi : 10.1007 / BF02172034 .
- ↑ Holwerda BC, Wittwer AJ, Duffin KL, Smith C, Toth MV, Carr LS, Wiegand RC, Bryant ML: Activity of two-chain recombinant human cytomegalovirus protease . In: J. Biol. Chem. . 269, No. 41, October 1994, pp. 25911-25915. PMID 7929296 .
- ↑ Loveland AN, Chan CK, Brignole EJ, Gibson W: Cleavage of human cytomegalovirus protease pUL80a at internal and cryptic sites is not essential but enhances infectivity . In: J. Virol. . 79, No. 20, October 2005, pp. 12961-12968. doi : 10.1128 / JVI.79.20.12961-12968.2005 . PMID 16188998 . PMC PMC1235863 (free full text).
- ↑ Pray TR, Nomura AM, Pennington MW, Craik CS: Auto-inactivation by cleavage within the dimer interface of Kaposi's sarcoma-associated herpesvirus protease . In: J. Mol. Biol. . 289, No. 2, June 1999, pp. 197-203. doi : 10.1006 / jmbi.1999.2791 . PMID 10366498 .
- ↑ Burck PJ, Berg DH, Luk TP, Sassmannshausen LM, Wakulchik M, Smith DP, Hsiung HM, Becker GW, Gibson W, Villarreal EC: Human cytomegalovirus maturational proteinase: expression in Escherichia coli, purification, and enzymatic characterization by using peptide substrate mimics of natural cleavage sites . In: J. Virol. . 68, No. 5, May 1994, pp. 2937-2946. PMID 8151764 . PMC PMC236782 (free full text).
- ↑ Shahian T, Lee GM, Lazic A, Arnold LA, Velusamy P, Roels CM, Guy RK, Craik CS: Inhibition of a viral enzyme by a small-molecule dimer disruptor . In: Nat. Chem. Biol . 5, No. 9, September 2009, pp. 640-646. doi : 10.1038 / nchembio.192 . PMID 19633659 . PMC PMC2752665 (free full text).
