Polygeny
Polygeny (derived from ancient Greek for multiple descent, English polygene mostly used as polygenic only as an adjective) is a term from genetics . It is used for cases in which the expression of a characteristic of the phenotype in the respective interest , for example a hereditary disease , depends on more than one single gene . If the observed distribution of traits can be explained by genetic differences, called different alleles , at a single gene locus , one speaks, in contrast, of monogeny .
Just as a single trait can very often depend on different genes, it is also common that a single gene (actually: an allele of a gene) influences different traits at the same time, this is then related to the gene pleiotropia or, related to the trait itself , Called polyphenia . Simplified it can therefore be said: polygeny: one characteristic, different genes; Polyphenia (or pleiotropy): one gene, different characteristics.
In classical formal genetics (also Mendelian genetics), inheritance patterns that are characterized by the interaction of different genes are difficult to investigate because the distribution of traits does not follow Mendel's third rule (independence rule ). If only a few genes are affected, the distribution of traits in cross-breeding experiments can usually be traced back to a larger, but manageable series of supplementary rules that take into account the form of these interactions. If - as with most real traits - many or very many genes are involved in the expression of the trait, this research approach fails completely. These characteristics are called “quantitative” (also complex or additive) characteristics. According to a research approach going back to Ronald Aylmer Fisher , their investigation is carried out with statistical methods, the research direction is called quantitative genetics . The investigation of polygenic traits roughly corresponds to the area of quantitative genetics, that of monogenic traits of Mendelian genetics. Traits that are influenced by a manageable number of genes fall into a gray area in which both approaches can be used in competition. Classically, these were also called polygenic, but today the expression “oligogenic” has become established instead. Today geneticists, in contrast to the pioneering days of genetics, speak of polygeny mostly only for quantitative characteristics that depend on a large (often not countable) number of genes.
Demarcation
Polygeny occurs when a single trait is influenced by multiple genes. This influence can manifest itself in different ways.
Epistasis
In his classic experiments on peas, Gregor Mendel found that the seven traits he was investigating (the growth form, the color and surface shape of the seeds and the flower color) in the pea were inherited independently of one another. For each of these traits he postulated a genetic disposition later called a gene, one sentence of which is passed on (inherited) from the father or mother to the offspring. The variability of the traits in his experiments, in which each trait occurred in two qualitatively different forms, was based on only one single gene per trait. Interactions between these genes did not occur or were at least negligibly small. Most of the characteristics of interest to researchers, and their genes, behave differently (so that it can be assumed that Mendel selected his test subjects carefully and after appropriate preliminary tests). Such interactions between genes are commonly called epistasis . (It is important to note: The expression was originally defined in Mendelian genetics by William Bateson exclusively for cases in which the influence of one gene locus on the phenotype was completely covered (masked) by the influence of a second; today it is also used for everyone used in other cases in which some relationship between two gene loci affects the phenotype.) If epistasis is present, the influence of a gene on the examined trait depends on the genetic polymorphism as well as on the presence of the allele under consideration at this gene locus (or several) other genes.
Coupling, haplotypes, linkage disequilibrium
If one looks at the inheritance of traits that are inherited polygenically, it often becomes apparent that certain traits occur together more often than would be expected with random, independent inheritance (as assumed by Mendel's third rule, for example). In genetics, this refers to the coupling of these characteristics or the genes that cause them. In the early days of genetics, by patiently logging many crossbreeding experiments, researchers were able to identify groups of genes that are often inherited as linked together. Such “coupling groups” usually correspond to the different chromosomes . Coupling can be broken by crossing-over during meiosis , in which homologous DNA segments are exchanged between paternal and maternal chromosomes of the diploid chromosome set. Considered for individual gene pairs, this happens only very rarely. Using linkage analysis , geneticists can examine the linkage based on the phenotypic distribution of traits. Today, however, in many cases unknown genes are assigned directly by means of known sequences stored in gene banks ; the technique is called genome-wide association study , abbreviated GWAS. Linking genes in the case of polygenically inherited traits has no effect at all on the individual phenotype. Only its distribution and frequency is different than expected.
Alleles of genes that are on the same chromosome are linked to one another and are usually inherited together, since in meiosis, only the haploid chromosome sets are redistributed without crossing-over effects. If, on the other hand, the alleles are on different chromosomes, their joint inheritance is only 50 percent likely. Because of this meaning, a new technical term was added for the genes that occur together: haplotype . In the case of the human genome, it is now known that large sections within a chromosome are almost always inherited in blocks; there is almost no crossing-over within the blocks. These haplotype blocks often keep haplotypes unchanged over many generations. Their analysis is of great importance, among other things for the unraveling of the individual lineage of individuals and populations.
The term linkage disequilibrium (roughly translated as “gene linkage imbalance”, but is usually no longer translated as a technical term) is based on the results of linkage analyzes. Today, however, this expression is used in a broader sense for all associations of different alleles in the genome with one another, the distribution of which deviates from the random distribution, regardless of whether this is due to coupling or not; the use of the term for it has only historical reasons. Linkage disequilibrium, i.e. the non-random occurrence of different alleles together, can be calculated within populations and between populations. It is initially independent of whether the sequence segments or genes linked in this way actually act on a certain characteristic together, i.e. whether there is polygeny. However, this is often the case and represents an important research approach to find out.
Linkage disequilibrium and epistasis have no direct factual relationship either; linkage disequilibrium only influences the frequency distribution of a polygenic trait in a population studied, not the genetic basis of this trait itself.
Additive polygeny
In additive polygeny, different genes work together in the formation of a characteristic and add up in their effect. If many genes are involved in the expression of the trait of interest, this results in a seamless sequence (no segregation of traits), for example in the case of body size not two, three or more discrete size classes, but a continuously varying spectrum from the smallest to the largest individual . This follows a Gaussian curve in the overall distribution . The total variance also results additively from the partial variances of the respective individual genes. Characteristics that correspond to this profile are called "quantitative" characteristics by geneticists. Most of the time, the term polygeny is used exclusively to refer to them today. Although the different alleles at the different gene loci involved in the expression of traits, each considered individually, can be characterized in their distribution following Mendel's rules, the overall result for many genes is a stepless and continuous distribution. Epistasis between individual genes does not normally change the distribution either, since the net effect of epistasis has the same effect in most contexts as a corresponding gene with an actual additive effect (“apparent” additive variance). In the case of quantitative traits, the clear connection between gene and trait is always lost. In the framework of formal genetics one speaks of incomplete penetrance , i.e. H. even with a dominant gene, not all individuals who are carriers of the corresponding allele show the phenotypic characteristic. In the case of diseases, the outbreak of the disease can no longer be predicted, but only a greater or lesser risk (medical: a disposition ) for it. In addition, the same phenotype (here: the disease) can be due to very different combinations of genes in different patients.
The individual components of additive polygeny are referred to using the English technical term Quantitative Trait Locus , usually abbreviated as QTL. Typically, however, the components found in the QTL analysis often do not add up to the overall heritability (heredity). What this discrepancy is based on is still scientifically controversial. A plausible answer here too would be an epistatic effect resulting from the interaction of gene networks, as studies on model organisms suggest.
Additive polygeny (the term goes back to English biometric researchers such as Kenneth Mather ) was known for a long time in German-speaking countries with the synonymous term polymer . Some differentiated further and called homomerism the additive effect of equally strong effects, with heteromerism (or polymer in the narrower sense) the interaction of factors of different strengths. The basis was primarily the classic experiments on the grain color of oats and wheat by the plant geneticist Herman Nilsson-Ehle (1873–1949) published in 1909
Polygeny and environmental influences (multifactorial inheritance)
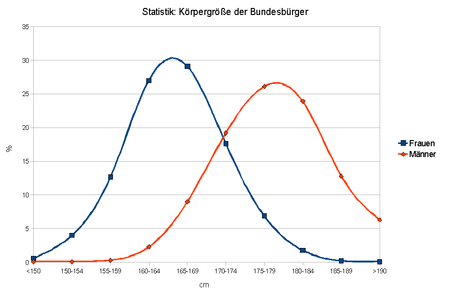
If a trait is dependent on several genes as well as on environmental factors, one speaks of multifactorial inheritance . Many geneticists want to limit the term polygenic inheritance to cases in which the characteristic variance is exclusively genetically determined, i.e. environmental influences should be excluded; however, this is often not taken into account in actual language usage.
When considering an entire population in terms of a quantitative characteristic, the interplay of polygeny and environmental factors leads to a continuous variance of the phenotype (within a certain framework). The more genes involved, the more continuous the curve becomes. The continuous variability (both individually influenced by polygeny or environmental factors, as well as resulting from the interaction of these) leads to a distribution pattern that corresponds to that of the Gaussian curve . Overall, many human characteristics (IQ, height, weight) follow the normal distribution .
- Examples
The skin color . Numerous, possibly hundreds, of genes are involved in the development of skin pigmentation in humans. This enables a very wide gradation between very dark and very light skin color. Environmental influences such as UV radiation also change the phenotype of the skin. The production of melanin in the skin is additionally stimulated within genetically determined limits and the skin appears darker. This reaction norm also has a genetic basis.
The body size . First of all, it is inherited to a certain extent. That is, big parents usually have big children. However, the size actually achieved also depends on the quality of human nutrition ; above all on the abundant intake of protein. However, the individual genetic constitution limits the size to a maximum.
Complementary polygeny
In a classic experiment in 1905, William Bateson and Reginald Punnett found that when two pure white flowering varieties of the sweet pea ( Lathyrus odoratus ) were crossed, violet flowering offspring also occurred. The segregation of the characteristics deviates from the ratios to be expected according to Mendel's third rule. They were able to show that the result can be explained by the interaction of two genes, in which the characteristic only occurs if a certain dominant allele is pronounced in both genes. If the alleles A and B of the two genes code for purple flowers and their mutants a and b for white flowers, violet is only expressed if at least one allele A and one B are present in the genome (combination A_B_, e.g. AaBb , AABb, AaBB etc.). In the original varieties, the expression AAbb and aaBB can be reconstructed, both of which produce white flowers. If you look at all possible combinations, a ratio of 9 to 7 results for the purple flowered variant, i.e. H. seven combinations result in purple, nine white flowers. For those conditions in which a phenotype depends on the interaction of two genes that complement each other, the term complementary polygeny was introduced. Today we know that the genetic basis for this is usually chains or cascades of genes connected one behind the other, which fail if any link in the chain fails. After the " Dobzhansky - Muller model" (actually first proposed by Bateson) complementary genes may play an important role in speciation play in which they the otherwise implausible, origin of the sterility of hybrids allow between newly formed species.
Complementary genes are now considered an example of epistasis. In contrast to the earlier usage, they are no longer included under the term polygeny. The earlier usage can still be found in many older works, including school books and exam papers.
Individual evidence
- ↑ R. Rieger, A. Michaelis, MM Green: Glossary of Genetics and Cytogenetics: Classical and Molecular. Springer Verlag, 4th edition 2012. ISBN 978-3-642-96327-8 . on page 74.
- ↑ cf. “Polygene: one of a group of genes that together control a quantitative character.” (One of a group of genes that together control a quantitative trait). in: Robert C. King, William D. Stansfield, Pamela K. Mulligan: A Dictionary of Genetics. Oxford University Press, seventh edition 2006. ISBN 978-0-19-530762-7 .
- ↑ on the molecular identity of the classical Mendelian genes cf. James B. Reid & John J. Ross (2011): Mendel's Genes: Toward a Full Molecular Characterization. Genetics 189 (1): 3-10. doi: 10.1534 / genetics.111.132118
- ↑ a b Trudy FC Mackay (2014): Epistasis and quantitative traits: using model organisms to study gene – gene interactions. Nature Reviews Genetics 15: 22-33. doi: 10.1038 / nrg3627
- ↑ Jurg Ott, Jing Wang, Suzanne M. Leal (2015): Genetic linkage analysis in the age of whole-genome sequencing. Nature Reviews Genetics 16 (5): 275-284. doi: 10.1038 / nrg3908
- ↑ Montgomery Slatkin (2008): Linkage disequilibrium - understanding the evolutionary past and mapping the medical future. Nature Reviews Genetics 9: 477-485. doi: 10.1038 / nrg2361
- ^ Julius Bauer: Lectures on general constitution and inheritance for students and doctors. Julius Springer Verlag, Berlin, second edition 1923. therein sixth lecture: The experimental-biological (Mendelian) laws of inheritance.
- ↑ Margarete Weninger (1976): On polygenic and multifactorial inheritance. Anthropologischer Anzeiger 35 (4): 236-239. JSTOR 29538872
- ↑ Jochen Graw, Wolfgang Hennig: Genetics. 5th, completely revised. Edition. Springer, Berlin / Heidelberg 2010, ISBN 978-3-642-04998-9 , pp. 456–459.
- ↑ Multifactorial (polygenic) inheritance. In: Werner Buselmaier, Gholamali Tariverdian: Human Genetics. 3rd, update and edit again Edition. Springer, 2004, ISBN 3-540-00873-X , p. 226.
- ↑ Robert Elston, Jaya Satagopan, Shuying Sun (2012): Genetic Terminology. Methods of Molecular Biology 850: 1-9. doi : 10.1007 / 978-1-61779-555-8_1
- ^ Arnold Lohaus, Marc Vierhaus, Asja Maass: Developmental Psychology of Childhood and Adolescence for Bachelor. 1st edition. Springer, Berlin / Heidelberg 2010, ISBN 978-3-642-03935-5 , p. 126.
- ^ AK Kalla (2007): Human Skin Color, Its Genetics, Variation and Adaptation: A Review. Anthropologist Special Issue No. 3: 209-214. PDF
- ↑ Genetics and Immunobiology. (Natura). Klett, Stuttgart 1997, ISBN 3-12-042939-2 . (Teachers' volume)
- ↑ cf. YS Demin: Mendel's Laws. FAO Corporate Document Repository , accessed on June 17, 2017.
- ↑ Patrick C. Phillips (2008): Epistasis - the essential role of gene interactions in the structure and evolution of genetic systems. Nature Reviews Genetics 9: 855-867. doi: 10.1038 / nrg2452
- ↑ H. Allen Orr (1996): Dobzhansky, Bateson and the Genetics of Speciation. Genetics 144: 1331-1335.
literature
- Jochen Graw, Wolfgang Hennig: Genetics. 5th edition. completely revised Edition. Springer, Berlin / Heidelberg 2010, ISBN 978-3-642-04998-9 , pp. 456–459.
- Multifactorial (polygenic) inheritance. In: Werner Buselmaier, Gholamali Tariverdian: Human Genetics. 3rd, update and edit again Edition. Springer, 2004, ISBN 3-540-00873-X , chapter 6.
Web links
- Polygenie - Article in human genetics at zum.de
- Entry Polygenie in the Lexicon of Biology , www.spektrum.de