Most recent common ancestor
Most Recent Common Ancestor or Last Common Ancestor ( English for the last common ancestor or also the youngest common ancestor ), abbreviated MRCA or LCA , is a technical term from the life sciences that is used in connection with researching the relationships between individuals or taxa .
Phylogenetics
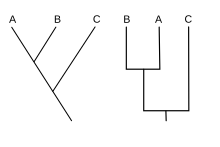
In the kladistisch embossed phylogenetics refers most recent common ancestor of the last common ancestor of two sister groups (Adelphotaxa). Ancestor is here not in the literal sense for an individual but for a hypothetical taxon in the species rank , the so-called ancestral species * . In a cladogram an ancestral species corresponds to the point, usually nodes called, in which a trunk line (line of development) by a Artbildungs event in two sister lines divides ( dichotomous branching ** ). A taxon whose sub-taxa can all be traced back to an MRCA is monophyletic or forms a clade . In the cladistic, modern biological system, only such taxa are recognized as valid in principle. A taxon whose sub-taxa do not have an MRCA is para- or polyphyletic . Special cases of the MRCA in cladistic sense are the load-universal common ancestor ( forefather , literally, the last universal common ancestor ), abbreviated LUCA , and the last eukaryotic common ancestor (literally last eukaryotic common ancestor ), LECA abbreviated. LUCA is the ancestral species of all living things today ( prokaryotes , protists , fungi , plants and animals ), as well as probably all extinct multicellular organisms. He was a primitive microbes art, the bacteria or a more original form of life (cf.. Ribozyt ) belongs, and the first two emerged from his species were also microbes. LECA, on the other hand, is the evolutionarily much younger progeny of all organisms with complex cells that have a “real” nucleus surrounded by a nuclear membrane, that is, it is at the base of that part of the “tree of life” that does not contain any prokaryotes.

Cladograms are the usual form of representation of the results of a cladistic relationship analysis, which today is usually carried out with the help of a specially developed computer program. The database required for this can be obtained both by recording morphological characteristics and their characteristic states and by examining the genetic material (mostly individual, selected genes or DNA sequences of nuclear or mitochondrial DNA ) of exemplary representatives of those species whose relationship is to be researched. The resulting cladogram can then easily be read off which species and higher-level taxa go back to a most recent common ancestor and which do not. Among other things, statements can be made about whether a certain characteristic that occurs in several different species has been adopted by an MRCA, making it homologous , or whether it is independent - convergent - in one, several or even all of the species examined - was created. In addition, in morphologically based analyzes, each node has a certain combination of feature states , which are the original ( ancestral ) feature states for the respective clade . These correspond to the feature states in the MRCA of this clade. Analogous to this, gene-based analyzes provide the ancestral base pair configurations of the examined DNA sequence for a clade. In addition, based on phylogenetic hypotheses determined elsewhere , ancestral states of this gene can be reconstructed with the aid of special computer programs from a data set of DNA sequences that encode a certain protein . The corresponding ancestral proteins can then be synthesized ( expressed ) in vitro and their properties can be examined.
With the help of the molecular clock , the point in time § in the past at which two lines of development separated (English: divergence date ) can be determined from a molecular genetic data set, i.e. the point in time at which the common parent species of a certain clade split into two new styles split. When determining the times of divergence of as many subclades as possible in a clade , the time course of adaptive radiations in this clade can be determined and conclusions can be drawn about distribution paths (i.e. the paleobiogeography).
Population Genetics and Genetic Genealogy
In population genetics , Most Recent Common Ancestor refers to an individual who is the last common ancestor of a group of individuals living today. The term is also used in the same sense in genetic genealogy , although in this discipline the investigations are largely limited to humans.
In population genetics and genetic genealogy, the MCRA is usually determined using DNA sequences of the Y chromosome or mitochondrial DNA. More precisely, similar to cladistic analyzes on a molecular basis, the base pair configuration of the MRCA of this sample is reconstructed from the genetic data of a suitable sample by means of an algorithm for the DNA sequence in question. On this basis, for example, it can then be determined in which existing population the genetic agreement with this MRCA is greatest. The geographic region in which the population with the closest match lives can then be considered the likely region of origin of the MRCA. So u. a. find out in which refuges widespread species in Europe survived the last ice age . The so-called Out-of-Africa-II theory about the origin and distribution of modern humans ( Homo sapiens ) is also supported by the results of a comparative study of DNA sequences of human mitochondrial DNA (see also Mitochondrial Eva ).
In addition, the reconstructed base pair configuration of an MRCA can also be used in population genetics and genetic genealogy as a basis for the use of the molecular clock, which can be used to determine when the MRCA in question lived in the past.
Remarks
- * Richard Dawkins used in his popular science book The Ancestor's Tale , which describes a journey along the human lineage, for MRCAS of different inclusive clades to which man belongs ( primates , mammals , amniotes , etc.), the expression concestor (in the German version translated as "fellow passenger").
- ** In cladistics, a parent species is always split into two new species by convention
- § The word "time" in this context means strictly speaking, for a time space , as viewed in human time scales, speciation is more of an ongoing process over several generations as a single event. In relation to the large periods of time in the history of the earth and in the evolutionary history of a relatively inclusive clade, this period can be understood as a point in time and the speciation as a single event.
Individual evidence
- ↑ Joran Martijn, Thijs JG Ettema: From archaeon to eukaryote: the evolutionary dark ages of the eukaryotic cell. Biochemical Society Transactions. Vol. 41, No. 1, 2013, pp. 451-457, doi: 10.1042 / BST20120292
- ↑ z. B. the ancestral rhodopsin of the amniotes, mammals (Mammalia) and higher mammals (Theria), see Constanze Bickelmann, James M. Morrow, Jing Du, Ryan K. Schott, Ilke van Hazel, Steve Lim, Johannes Müller, Belinda SW Chang: The molecular origin and evolution of dim-light vision in mammals. Evolution. Vol. 69, No. 11, 2015, pp. 2995-3003, doi: 10.1111 / evo.12794 ; see also Constanze Bickelmann: Visual pigment evolution and the paleobiology of early mammals. Dissertation to obtain the academic degree doctor rerum naturalium (Dr. rer. Nat.) In biology, Faculty of Mathematics and Natural Sciences at the Humboldt University of Berlin, 2011, urn : nbn: de: kobv: 11-100191633
- ↑ see e.g. B. a work on the phylogeny of the plant family Rapateaceae : TJ Givnish, TM Evans, ML Zjhra, TB Patterson, PE Berry, KJ Sytsma: Molecular evolution, adaptive radiation, and geographic diversification in the amphiatlantic family Rapateaceae: evidence from ndh F sequences and morphology. Evolution. Vol. 54, No. 6, 2000, pp. 1915-1937, doi: 10.1111 / j.0014-3820.2000.tb01237.x
- ↑ a b Bruce Walsh: Estimating the time to the most recent common ancestor for the Y chromosome or mitochondrial DNA for a pair of individuals. Genetics. Vol. 158, No. 2, 2001, pp. 897−912 ( PDF ).
- ↑ for example the chaffinch , see Cortland K. Griswold, Allan J. Baker: Time to the most recent common ancestor and divergence times of populations of common chaffinches ( Fringilla coelebs ) in Europe and North Africa: insights into Pleistocene refugia and current levels of migration. Evolution. Vol. 56, No. 1, 2002, pp. 143-153, doi : 10.1111 / j.0014-3820.2002.tb00856.x .
- ↑ Richard Dawkins: Stories from the Origin of Life. Ullstein, Berlin 2008, footnote p. 22, ISBN 978-3-550-08748-6
