Wikipedia talk:Userboxes/Sports and Chromosome: Difference between pages
m Reverted edits by 207.176.229.1 to last version by Darth Panda (HG) |
|||
| Line 1: | Line 1: | ||
{{for|information about chromosomes in [[genetic algorithm]]s|chromosome (genetic algorithm)}} |
|||
{{talkheader}} |
|||
{{for|a non-technical introduction to the topic|Introduction to genetics}} |
|||
{{WikiProject Userboxes}} |
|||
[[Image:Chromosome-upright.png|frame|Diagram of a duplicated and condensed ([[metaphase]]) eukaryotic chromosome. (1) [[Chromatid]] - one of the two identical parts of the chromosome after [[S phase]]. (2) [[Centromere]] - the point where the two chromatids touch, and where the microtubules attach. (3) Short arm. (4) Long arm.]] |
|||
__TOC__ |
|||
'''Chromosomes''' are organized structures of [[DNA]] and [[protein]]s that are found in [[Cell (biology)|cells]]. A chromosome is a singular piece of DNA, which contains many [[gene]]s, [[regulatory sequence|regulatory elements]] and other [[genetic sequence|nucleotide sequences]]. Chromosomes also contain DNA-bound proteins, which serve to package the DNA and control its functions. The word ''chromosome'' comes from the [[Greek language|Greek]] {{polytonic|χρῶμα}} (''chroma'', color) and {{polytonic|σῶμα}} (''soma'', body) due to their property of being stained very strongly by some [[dyes]]. |
|||
For sports users play how about running/track?--[[User:CHIMES|CHIMES]] 15:32, 1 January 2006 (UTC) |
|||
Chromosomes vary extensively between different [[organism]]s. The DNA molecule may be circular or linear, and can contain anything from tens of [[kilobase]] pairs to hundreds of [[megabase]] pairs. Typically [[eukaryote|eukaryotic]] cells (cells with nuclei) have large linear chromosomes and [[prokaryotic]] cells (cells without defined nuclei) have smaller circular chromosomes, although there are many exceptions to this rule. Furthermore, cells may contain more than one type of chromosome; for example, [[mitochondria]] in most [[eukaryotes]] and [[chloroplasts]] in plants have their own small chromosomes. |
|||
In eukaryotes, nuclear chromosomes are packaged by proteins into a condensed structure called [[chromatin]]. This allows the very long DNA molecules to fit into the [[cell nucleus]]. The structure of chromosomes and chromatin varies through the [[cell cycle]]. Chromosomes may exist as either duplicated or unduplicated—unduplicated chromosomes are single linear strands, whereas duplicated chromosomes (copied during [[S phase|synthesis phase]]) contain two copies joined by a [[centromere]]. Compaction of the duplicated chromosomes during [[mitosis]] and [[meiosis]] results in the classic four-arm structure (pictured to the right). |
|||
== Boxing == |
|||
There is no boxing userbox??--[[User:GorillazFanAdam|GorillazFanAdam]] 03:40, 13 April 2006 (UTC) |
|||
"Chromosome" is a rather loosely defined term. In prokaryotes, a small circular DNA molecule may be called either a [[plasmid]] or a small chromosome. These small circular genomes are also found in mitochondria and chloroplasts, reflecting their bacterial origins. The simplest chromosomes are found in [[viruses]]: these DNA or RNA molecules are short linear or circular chromosomes that often lack any structural proteins. |
|||
== |
==History== |
||
This is a brief history of research in a complex field where each advance was hard won, and often hotly disputed at the time. |
|||
'''Visual discovery of chromosomes'''. Textbooks have often said that chromosomes were first observed in [[plant]] cells by a [[Switzerland|Swiss]] [[botanist]] named [[Karl Wilhelm von Nägeli]] in 1842.<ref>Nägeli C. 1842. ''Zur Entwickelungsgeschichte des Pollens bei den Phanerogamen''. Ovell & Füssli, Zürich.</ref> However, this opinion has been challenged, perhaps decisively, by [[Henry Harris (scientist)|Henry Harris]], who has freshly reviewed the primary literature.<ref>Harris H. 1999. ''The birth of the cell''. Yale University Press. p138</ref> In his opinion the claim of Nägeli to have seen spore mother cells divide is mistaken, as are some of his interpretations. Harris considers other candidates, especially [[Wilhelm Hofmeister]], whose publications in 1848-9 include plates that definitely show mitotic events.<ref>Hofmeister W. 1848. ''Bot. Zeit''. '''6''', cols 425, 649, 670.</ref><ref>Hofmeister W. 1849. ''Die Entstehung des Embryos der Phanerogamen''. Friedrich Hofmeister, Leipzig.</ref> Hofmeister was also the choice of [[Cyril Darlington]]. |
|||
We need a Tae Kwon Do template! --[[User:Marudubshinki|maru]] [[User talk:Marudubshinki|(talk)]] [[Special:Contributions/Marudubshinki|Contribs]] 02:25, 2 January 2006 (UTC) |
|||
The work of other cytologists such as [[Walther Flemming]], [[Eduard Strasburger]], [[Otto Bütschli]], [[Oskar Hertwig]] and [[Carl Rabl]] should be acknowledged. The use of [[basophilic]] [[aniline]] [[dye]]s was a new technique for effectively [[staining]] the [[chromatin]] material in the nucleus. Their behavior in animal ([[salamander]]) cells was later described in detail by [[Walther Flemming]], who in 1882 "provided a superb summary of the state of the field."<ref>Mayr E. 1982. ''The growth of biological thought''. Harvard. p677</ref><ref>Flemming W. ''Zellsubstanz Kern und Zelltheilung''. Vogel, Leipzig.</ref> The name ''chromosome'' was invented in 1888 by [[Heinrich Wilhelm Gottfried von Waldeyer-Hartz|Heinrich von Waldeyer]]. However, van Beneden's monograph of 1883 on the fertilised eggs of the parasitic roundworm ''[[Ascaris]] megalocephala'' was the outstanding work of this period.<ref>Van Beneden E. 1883. ''Arch. Biol.'' '''4'''.</ref> His conclusions are classic: |
|||
Ok, I agree. We got Judo and we got Karate. Where's Taekwondo?! [[User:Spyco|Spyco]] 05:08, 21 May 2006 (UTC) |
|||
*Thus there is no fusion between the male chromatin and the female chromatin at any stage of division.... |
|||
*The elements of male origin and those of female origin are never fused together in a cleavage nucleus, and perhaps they remain distinct in all the nuclei derived from them. [tranl: Harris p162] |
|||
<blockquote>"It is not easy to identify who first discerned chromosomes during mitosis, but there is no doubt that those who first saw them had no idea of their significance... [but] with the work of [[Edouard-Gérard Balbiani|Balbiani]] and [[Edouard Van Beneden|van Beneden]] we move away from... the mechanism of cell division to a precise delineation of chromosomes and what they do during the division of the cell."<ref>Harris H. 1999. ''The birth of the cell''. Yale University Press. p141, 153</ref></blockquote> |
|||
Here you go :) Although there are belts and a federation, I thought some users may want a simple "taekwondo practitioner" box with a category already included. [[User:Pundit|<span style="color: blue;">Pundit</span>]]|[[User talk:Pundit|<span style="color: green;">utter</span>]] 00:39, 27 December 2007 (UTC) |
|||
Van Beneden's master work was closely followed by that of Carl Rabl, who reached similar conclusions. <ref>Rabl C. 1885. ''Morphol. Jahrb''. '''10''', 24.</ref> This more or less concludes the first period, in which chromosomes were visually sighted and the morphological stages of mitosis were described. Coleman also gives a useful review of these discoveries.<ref>Coleman W. 1965. Cell, nucleus, and inheritance: an historical discovery. ''Proc. Am. Philos. Soc''. '''109''', 124-158.</ref> |
|||
{{Template:Taekwondo}} {{clear}} |
|||
<nowiki>{{Template:Taekwondo}}</nowiki> |
|||
'''Nucleus as the seat of heredity'''. The origin of this epoch-making idea lies in a few sentences tucked away in Ernst Haeckel's ''Generelle Morphologie'' of 1866.<ref>Haeckel E. 1866. ''Generelle Morphologie der Organismen: Allgemeine Gründzuge der organischen Formen-Wissenschaft''. 2 vols, Reimer, Berlin.</ref> The evidence for this insight gradually accumulated until, after twenty or so years, two of the greatest in a line of great German scientists spelled out the concept. [[August Weismann]] proposed that the [[germ line]] is separate from the [[somatic cell|soma]], and that the cell nucleus is the repository of the hereditary material, which, he proposed, is arranged along the chromosomes in a linear manner. Furthermore, he proposed that at fertilisation a new combination of chromosomes (and their hereditary material) would be formed. This was the explanation for the reduction division of [[meiosis]] (first described by van Beneden). |
|||
== Suggestions == |
|||
I did these up. Feel free to use them. [[User:Trekphiler|Trekphiler]] 13:28, 2 January 2006 (UTC) |
|||
{{Userbox |
|||
|border-c=black |
|||
|id-c=white |
|||
|id-s=8 |
|||
|id-fc=black |
|||
|info-c=white |
|||
|info-s=8 |
|||
|info-fc=black |
|||
|id=[[Image:Karthikeyan (Jordan) locking brakes in qualifying at USGP 2005.jpg|50px]] |
|||
|info=This user follows [[Formula 3000]]. }} |
|||
{{Userbox |
|||
|border-c=black |
|||
|id-c=red |
|||
|id-s=8 |
|||
|id-fc=black |
|||
|info-c=white |
|||
|info-s=8 |
|||
|info-fc=black |
|||
|id=[[Image:top_fuel_eg.jpg|50px]] |
|||
|info=This user enjoys [[drag racing]]. |
|||
}} |
|||
{{Userbox |
|||
|border-c=black |
|||
|id-c=white |
|||
|id-s=8 |
|||
|id-fc=black |
|||
|info-c=white |
|||
|info-s=8 |
|||
|info-fc=black |
|||
|id=[[Image:Karthikeyan (Jordan) locking brakes in qualifying at USGP 2005.jpg|50px]] |
|||
|info=This user follows [[GP2]]. }} |
|||
{{userbox|red|white|[[Image:Honda_beat.jpg|60px]]|This user wishes [[Department of Transportation|DOT]] allowed the [[Honda Beat]] to be imported.}} |
|||
{{Userbox |
|||
|border-c=black |
|||
|id-c=white |
|||
|id-s=8 |
|||
|id-fc=black |
|||
|info-c=white |
|||
|info-s=8 |
|||
|info-fc=black |
|||
|id=[[Image:Cowboy.jpg|50px]] |
|||
|info=This user supports [[Cowboy action shooting ]]. }} |
|||
{{Userbox |
|||
|border-c=black |
|||
|id-c=yellow |
|||
|id-s=8 |
|||
|id-fc=black |
|||
|info-c=white |
|||
|info-s=8 |
|||
|info-fc=black |
|||
|id=[[Image:Pistolet-marine-19e-1.png|50px]] |
|||
|info=This user supports [[practical pistol]] shooting. }} |
|||
<br><br><br><br> |
|||
<br><br><br><br> |
|||
I did this up. Feel free... 07:12, 20 June 2006 (UTC) |
|||
{{Userbox |
|||
|border-c=black |
|||
|id-c=white |
|||
|id-s=8 |
|||
|id-fc=black |
|||
|info-c=white |
|||
|info-s=8 |
|||
|info-fc=black |
|||
|id=[[Image:Karthikeyan (Jordan) locking brakes in qualifying at USGP 2005.jpg|50px]] |
|||
|info=This user follows [[GP2]]. }} |
|||
<br><br><br><br> |
|||
---- |
|||
I did this up. Feel free... [[User:Trekphiler|Trekphiler]] 04:10, 6 January 2006 (UTC) |
|||
{{userbox|green|white|[[Image:Ccross.svg|40px]]|This user salutes [[Boston]] for trying to keep white guys in the [[NBA]].}} |
|||
'''Chromosomes as vectors of heredity'''. In a series of outstanding experiments, [[Theodor Boveri]] gave the definitive demonstration that chromosomes are the vectors of heredity. His two principles were: |
|||
<br><br><br><br> |
|||
:The continuity of chromosomes |
|||
---- |
|||
:The individuality of chromosomes. |
|||
It is the second of these principles that was so original<!--how precise language can be when used properly-->. Boveri was able to test the proposal put forward by [[Wilhelm Roux]], that each chromosome carries a different genetic load, and showed that Roux was right. Upon the rediscovery of Mendel, Boveri was able to point out the connection between the rules of inheritance and the behaviour of the chromosomes. It is interesting to see that Boveri influenced two generations of American cytologists: [[Edmund Beecher Wilson]], [[Walter Sutton]] and [[Theophilus Painter]] were all influenced by Boveri (Wilson and Painter actually worked with him). In his famous textbook ''The Cell'', Wilson linked Boveri and Sutton together by the [[Boveri-Sutton Chromosome Theory|Boveri-Sutton theory]]. Mayr remarks that the theory was hotly contested by some famous geneticists: [[William Bateson]], [[Wilhelm Johannsen]], [[Richard Goldschmidt]] and [[T.H. Morgan]], all of a rather dogmatic turn-of-mind. Eventually complete proof came from chromosome maps – in Morgan's own lab.<ref>Mayr E. 1982. ''The growth of biological thought''. Harvard. p749</ref> |
|||
What about userboxes for referees/officials? [[User:Amelia9mm|Amelia9mm]] <small>—Preceding [[Wikipedia:Signatures|comment]] was added at 03:45, 10 February 2008 (UTC)</small><!--Template:Undated--> <!--Autosigned by SineBot--> |
|||
<br><br><br><br> |
|||
==Chromosomes in eukaryotes== |
|||
== Badminton == |
|||
{{Mergefrom|Eukaryotic chromosome fine structure|date=December 2007}} |
|||
[[Eukaryotes]] ([[cell (biology)|cells]] with nuclei such as plants, yeast, and animals) possess multiple large linear chromosomes contained in the cell's nucleus. Each chromosome has one [[centromere]], with one or two arms projecting from the centromere, although, under most circumstances, these arms are not visible as such. In addition, most eukaryotes have a small circular [[mitochondria]]l genome, and some eukaryotes may have additional small circular or linear [[cytoplasm]]ic chromosomes. |
|||
In the nuclear chromosomes of [[eukaryote]]s, the uncondensed DNA exists in a semi-ordered structure, where it is wrapped around [[histone]]s (structural [[protein]]s), forming a composite material called [[chromatin]]. |
|||
I'd like to see a badminton userbox. - [[User:RoyBoy|Roy]][[User talk:RoyBoy|'''Boy''']] <sup>[[User:RoyBoy/The 800 Club|800]]</sup> 01:28, 4 February 2006 (UTC) |
|||
===Chromatin=== |
|||
{{Main|Chromatin}} |
|||
[[Image:Chromatin Structures.png|thumb|right|350px|'''Fig. 2:''' The major structures in DNA compaction; [[DNA]], the [[nucleosome]], the 10nm "beads-on-a-string" fibre, the 30nm fibre and the [[metaphase]] chromosome.]] |
|||
Chromatin is the complex of DNA and protein found in the [[eukaryote|eukaryotic]] nucleus which packages chromosomes. The structure of chromatin varies significantly between different stages of the [[cell cycle]], according to the requirements of the DNA. |
|||
====Interphase chromatin==== |
|||
== lax, baby == |
|||
During [[interphase]] (the period of the [[cell cycle]] where the cell is not dividing) two types of [[chromatin]] can be distinguished: |
|||
Did this one...although I'm not sure there are many lacrosse players on Wikipedia. [[User:Osbus|Osbus]] 00:50, 28 March 2006 (UTC) |
|||
* [[Euchromatin]], which consists of DNA that is active, e.g., expressed as protein. |
|||
{{User:UBX/Lax}} |
|||
* [[Heterochromatin]], which consists of mostly inactive DNA. It seems to serve structural purposes during the chromosomal stages. Heterochromatin can be further distinguished into two types: |
|||
** ''Constitutive heterochromatin'', which is never expressed. It is located around the centromere and usually contains [[Repeated sequence (DNA)|repetitive sequences]]. |
|||
** ''Facultative heterochromatin'', which is sometimes expressed. |
|||
Individual chromosomes cannot be distinguished at this stage - they appear in the nucleus as a homogeneous tangled mix of DNA and protein. |
|||
<br><br><br><br><br> |
|||
=== |
====Metaphase chromatin and division==== |
||
{{seealso|mitosis|meiosis}} |
|||
<!-- too many pictures! [[Image:ChickenChromosomesBMC Genomics5-56Fig4.jpg|thumb|200px|right|Chicken chromosomes during [[metaphase]], with highlighted [[locus (genetics)|genetic loci]]]] --> |
|||
[[Image:HumanChromosomesChromomycinA3.jpg|thumb|200px|right|Human chromosomes during [[metaphase]].]] |
|||
In the early stages of mitosis or meiosis (cell division), the chromatin strands become more and more condensed. They cease to function as accessible genetic material ([[Transcription (genetics)|transcription]] stops) and become a compact transportable form. This compact form makes the individual chromosomes visible, and they form the classic four arm structure, a pair of sister [[chromatids]] attached to each other at the [[centromere]]. The shorter arms are called ''p arms'' (from the [[French language|French]] ''petit'', small) and the longer arms are called ''q arms'' (''q'' follows ''p'' in the Latin alphabet). This is the only natural context in which individual chromosomes are visible with an optical [[microscope]]. |
|||
During divisions long [[microtubule]]s attach to the centromere and the two opposite ends of the cell. The microtubules then pull the chromatids apart, so that each daughter cell inherits one set of chromatids. Once the cells have divided, the chromatids are uncoiled and can function again as chromatin. In spite of their appearance, chromosomes are structurally highly condensed, which enables these giant DNA structures to be contained within a cell nucleus (Fig. 2). |
|||
{{user SF2016}} |
|||
Feel free to use. - [[User:Cribananda|Cribananda]] 02:17, 14 June 2006 (UTC) |
|||
<br><br><br> |
|||
=== Help? === |
|||
{{User:UBX/snooker}} |
|||
Like the pic, but I only watch (when TSN will actually broadcast it, the slimeballs!), but not play. Can somebody do a fanbox? 07:08, 20 June 2006 (UTC) |
|||
The self-assembled microtubules form the spindle, which attaches to chromosomes at specialized structures called kinetochores, one of which is present on each sister [[chromatid]]. A special DNA base sequence in the region of the kinetochores provides, along with special proteins, longer-lasting attachment in this region. |
|||
== Added Navigation == |
|||
==Chromosomes in prokaryotes== |
|||
I just added the "WikiProject Userboxes" box at the bottom that seemed to be on every other section... |
|||
The prokaryotes - [[bacteria]] and [[archaea]] - typically have a single circular chromosome, but many variations do exist.<ref>{{cite journal |author=Thanbichler M, Shapiro L |title=Chromosome organization and segregation in bacteria |journal=J. Struct. Biol. |volume=156 |issue=2 |pages=292–303 |year=2006 |pmid=16860572 |doi=10.1016/j.jsb.2006.05.007}}</ref> Most bacteria have a single circular chromosome that can range in size from only 160,000 [[base pair]]s in the [[endosymbiont|endosymbiotic]] bacteria ''[[Candidatus Carsonella ruddii]]'',<ref>{{cite journal |author=Nakabachi A, Yamashita A, Toh H, Ishikawa H, Dunbar H, Moran N, Hattori M |title=The 160-kilobase genome of the bacterial endosymbiont Carsonella |journal=Science |volume=314 |issue=5797 |pages=267 |year=2006 |pmid=17038615 |doi=10.1126/science.1134196}}</ref> to 12,200,000 base pairs in the soil-dwelling bacteria ''[[Sorangium cellulosum]]''.<ref>{{cite journal |author=Pradella S, Hans A, Spröer C, Reichenbach H, Gerth K, Beyer S |title=Characterisation, genome size and genetic manipulation of the myxobacterium Sorangium cellulosum So ce56 |journal=Arch Microbiol |volume=178 |issue=6 |pages=484–92 |year=2002 |pmid=12420170 | doi = 10.1007/s00203-002-0479-2 <!--Retrieved from PMID by DOI bot-->}}</ref> [[Spirochaete]]s of the [[genus]] ''Borrelia'' are a notable exception to this arrangement, with bacteria such as ''[[Borrelia burgdorferi]]'', the cause of [[Lyme disease]], containing a single linear chromosome.<ref>{{cite journal |author=Hinnebusch J, Tilly K |title=Linear plasmids and chromosomes in bacteria |journal=Mol Microbiol |volume=10 |issue=5 |pages=917–22 |year=1993|pmid = 7934868 |doi=10.1111/j.1365-2958.1993.tb00963.x}}</ref> |
|||
===Structure in sequences=== |
|||
I'm new to Wikipedia, so if I did something incorrectly, please correct me :) |
|||
Prokaryotes chromosomes have less sequence-based structure than eukaryotes. Bacteria typically have a single point (the [[origin of replication]]) from which replication starts, whereas some archaea contain multiple replication origins.<ref>{{cite journal |author=Kelman LM, Kelman Z |title=Multiple origins of replication in archaea |journal=Trends Microbiol. |volume=12 |issue=9 |pages=399–401 |year=2004 |pmid=15337158 |doi=10.1016/j.tim.2004.07.001}}</ref> The genes in prokaryotes are often organized in [[operons]], and do not contain [[intron]]s, unlike eukaryotes. |
|||
===DNA packaging=== |
|||
[[User:FaerieInGrey|FaerieInGrey]] 13:22, 24 July 2006 (UTC) |
|||
[[Prokaryote]]s do not possess nuclei. Instead, their DNA is organized into a structure called the [[nucleoid]].<ref>{{cite journal |author=Thanbichler M, Wang SC, Shapiro L |title=The bacterial nucleoid: a highly organized and dynamic structure |journal=J. Cell. Biochem. |volume=96 |issue=3 |pages=506–21 |year=2005 |pmid=15988757 | doi = 10.1002/jcb.20519 <!--Retrieved from PMID by DOI bot-->}}</ref> The nucleoid is a distinct structure and occupies a defined region of the bacterial cell. This structure is, however, dynamic and is maintained and remodeled by the actions of a range of histone-like proteins, which associate with the bacterial chromosome.<ref>{{cite journal |author=Sandman K, Pereira SL, Reeve JN |title=Diversity of prokaryotic chromosomal proteins and the origin of the nucleosome |journal=Cell. Mol. Life Sci. |volume=54 |issue=12 |pages=1350–64 |year=1998 |pmid=9893710 |doi=10.1007/s000180050259}}</ref> In [[archaea]], the DNA in chromosomes is even more organized, with the DNA packaged within structures similar to eukaryotic nucleosomes.<ref>{{cite journal |author=Sandman K, Reeve JN |title=Structure and functional relationships of archaeal and eukaryal histones and nucleosomes | doi = 10.1007/s002039900122 <!--Retrieved from Yahoo! by DOI bot-->|journal=Arch. Microbiol. |volume=173 |issue=3 |pages=165–9 |year=2000 |pmid=10763747}}</ref><ref>{{cite journal |author=Pereira SL, Grayling RA, Lurz R, Reeve JN |title=Archaeal nucleosomes | doi = 10.1073/pnas.94.23.12633 <!--Retrieved from URL by DOI bot-->|journal=Proc. Natl. Acad. Sci. U.S.A. |volume=94 |issue=23 |pages=12633–7 |year=1997 |pmid=9356501 |url=http://www.pnas.org/cgi/pmidlookup?view=long&pmid=9356501}}</ref> |
|||
Bacterial chromosomes tend to be tethered to the [[plasma membrane]] of the bacteria. In molecular biology application, this allows for its isolation from plasmid DNA by centrifugation of lysed bacteria and pelleting of the membranes (and the attached DNA). |
|||
== Wrestling(WWE) == |
|||
Prokaryotic chromosomes and plasmids are, like eukaryotic DNA, generally [[Mechanical properties of DNA#Supercoiled DNA|supercoiled]]. The DNA must first be released into its relaxed state for access for [[Transcription (genetics)|transcription]], regulation, and [[DNA replication|replication]]. |
|||
Hey, I thought that there could be a few more WWE userboxes. I made these, feel free to use. |
|||
{{Userboxtop}} |
|||
{{userbox |
|||
|border-c=green |
|||
|info-c=black |
|||
|info-fc=green |
|||
|id-c=green |
|||
|id=[[Image:3085016.jpg|43px]] |
|||
|info=This user got told to [[D-Generation X|S*CK IT]]!}} |
|||
{{userbox |
|||
|border-c=black |
|||
|info-c=red |
|||
|info-fc=white |
|||
|id-c=white |
|||
|id=[[HBK]] |
|||
|info=This user [[Shawn Michaels|is a show stopper]]!}} |
|||
{{userbox |
|||
|border-c=blue |
|||
|info-c=black |
|||
|info-fc=white |
|||
|id-c=white |
|||
|id=[[RVD]] |
|||
|info=This user is the [[Rob Van Dam|WHOLE DAM SHOW]]!}} |
|||
{{userbox |
|||
|border-c=green |
|||
|info-c=black |
|||
|info-fc=white |
|||
|id-c=green |
|||
|id=[[Image:JohnCenaChain.JPG|43px]] |
|||
|info=This user is a [[John Cena|Chain Gang Soilder]]. Hustle, Loyalty, Respect!}} |
|||
{{userbox |
|||
|border-c=black |
|||
|info-c=white |
|||
|info-fc=black |
|||
|id-c=white |
|||
|id=[[Stone Cold|3:16]] |
|||
|info=This user believe's in [[Stone Cold|Austin 3:16]].}} |
|||
{{Userboxbottom}} |
|||
==Number of chromosomes in various organisms== |
|||
{{main|List of number of chromosomes of various organisms}} |
|||
===Eukaryotes=== |
|||
These tables give the total number of chromosomes (including sex chromosomes) in a cell nucleus. For example, human cells are [[diploid]] and have 22 different types of [[autosome]]s, each present as two copies, and two [[sex chromosomes]]. This gives 46 chromosomes in total. Other organisms have more than two copies of their chromosomes, such as [[Bread wheat]], which is ''hexaploid'' and has six copies of seven different chromosomes - 42 chromosomes in total. |
|||
{| style="float:right;"|} |
|||
== Where's the new XC-Skiing user box now? == |
|||
| |
|||
{| class="wikitable" style="margin:1em 0 1em 1em" |
|||
|+ Chromosome numbers in some plants |
|||
|- |
|||
! Plant Species !! # |
|||
|- |
|||
| ''[[Arabidopsis thaliana]]'' (diploid)<ref>{{cite journal |author=Armstrong SJ, Jones GH |title=Meiotic cytology and chromosome behaviour in wild-type Arabidopsis thaliana |journal=J. Exp. Bot. |volume=54 |issue=380 |pages=1–10 |year=2003 |month=January |pmid=12456750 |url=http://jexbot.oxfordjournals.org/cgi/pmidlookup?view=long&pmid=12456750 |doi=10.1093/jxb/54.380.1}}</ref> || 10 |
|||
|- |
|||
| [[Rye]] (diploid)<ref>{{cite journal |author=Gill BS, Kimber G |title=The Giemsa C-banded karyotype of rye |journal=Proc. Natl. Acad. Sci. U.S.A. |volume=71 |issue=4 |pages=1247–9 |year=1974 |month=April |pmid=4133848 |url=http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=388202&blobtype=pdf |doi=10.1073/pnas.71.4.1247}}</ref> || 14 |
|||
|- |
|||
| [[Maize]] (diploid)<ref>{{cite journal |author=Kato A, Lamb JC, Birchler JA |title=Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize |journal=Proc. Natl. Acad. Sci. U.S.A. |volume=101 |issue=37 |pages=13554–9 |year=2004 |month=September |pmid=15342909 |doi=10.1073/pnas.0403659101 |url=http://www.pubmedcentral.nih.gov/articlerender.fcgi?tool=pubmed&pubmedid=15342909}}</ref> || 20 |
|||
|- |
|||
| [[Einkorn wheat]] (diploid)<ref name=Dubcovsky>{{cite journal |author=Dubcovsky J, Luo MC, Zhong GY, ''et al'' |title=Genetic map of diploid wheat, Triticum monococcum L., and its comparison with maps of Hordeum vulgare L |journal=Genetics |volume=143 |issue=2 |pages=983–99 |year=1996 |pmid=8725244 |url=http://www.pubmedcentral.nih.gov/picrender.fcgi?artid=1207354&blobtype=pdf}}</ref> || 14 |
|||
|- |
|||
| [[Durum wheat]] (tetraploid)<ref name=Dubcovsky/> || 28 |
|||
|- |
|||
| [[Bread wheat]] (hexaploid)<ref name=Dubcovsky/> || 42 |
|||
|- |
|||
| [[Potato]] (tetraploid)<ref name=Ellison1935>{{cite journal | author = Ellison, W. | year = 1935 | title = A study of the chromosome numbers and morphology in certain British varieties of the common cultivated potato (solanum buberosum L.) | journal = Genetica | volume = 17 | issue = 1 | pages = 1–26 | url = http://www.springerlink.com/index/V8437QN6316363UX.pdf | accessdate = 2008-05-11 | doi = 10.1007/BF01984179}}</ref> || 48 |
|||
|- |
|||
| Cultivated tobacco (diploid)<ref>{{cite journal |author=Kenton A, Parokonny AS, Gleba YY, Bennett MD |title=Characterization of the Nicotiana tabacum L. genome by molecular cytogenetics |journal=Mol. Gen. Genet. |volume=240 |issue=2 |pages=159–69 |year=1993 |month=August |pmid=8355650 |doi=10.1007/BF00277053}}</ref> || 48 |
|||
|- |
|||
| [[Ophioglossum|Adder's Tongue Fern]] (diploid)<ref>{{cite journal |author=Leitch IJ, Soltis DE, Soltis PS, Bennett MD |title=Evolution of DNA amounts across land plants (embryophyta) |journal=Ann. Bot. |volume=95 |issue=1 |pages=207–17 |year=2005 |pmid=15596468 |url=http://aob.oxfordjournals.org/cgi/content/full/95/1/207 | doi = 10.1093/aob/mci014 <!--Retrieved from url by DOI bot-->}}</ref> || approx 1,440 |
|||
|} |
|||
| |
|||
{| class="wikitable" style="margin:1em 0 1em 1em" |
|||
|+ Chromosome numbers (2n) in some animals |
|||
|- |
|||
! Species !! # !! Species !! # |
|||
|- |
|||
| [[Drosophila melanogaster|Common fruit fly]] || 8 |
|||
| [[Guinea Pig]]<ref>{{cite journal |author=Umeko Semba, Yasuko Umeda, Yoko Shibuya, Hiroaki Okabe, Sumio Tanase and Tetsuro Yamamoto |title=Primary structures of guinea pig high- and low-molecular-weight kininogens | doi = 10.1016/j.intimp.2004.06.003 <!--Retrieved from URL by DOI bot-->|journal=International Immunopharmacology |volume=4 |issue=10-11 |pages=1391–1400 |year=2004 |url=http://www.sciencedirect.com/science/article/B6W7N-4CX6PRC-1/2/97487fdf611a04e3c88690da9e6d853b}}</ref>|| 64 |
|||
|- |
|||
| [[Dove]]{{Fact|date=February 2007}} || 16<!-- taxon? --> |
|||
| [[Garden snail]]<ref>{{cite journal |author=Vitturi R, Libertini A, Sineo L, ''et al'' |title=Cytogenetics of the land snails Cantareus aspersus and C. mazzullii (Mollusca: Gastropoda: Pulmonata) |journal=Micron |volume=36 |issue=4 |pages=351–7 |year=2005 |pmid=15857774 |doi=10.1016/j.micron.2004.12.010}}</ref> || 54 |
|||
|- |
|||
| [[Earthworm]] ''Octodrilus complanatus''<ref>{{cite journal |author=Vitturi R, Colomba MS, Pirrone AM, Mandrioli M |title=rDNA (18S-28S and 5S) colocalization and linkage between ribosomal genes and (TTAGGG)(n) telomeric sequence in the earthworm, Octodrilus complanatus (Annelida: Oligochaeta: Lumbricidae), revealed by single- and double-color FISH | doi = 10.1093/jhered/93.4.279 <!--Retrieved from URL by DOI bot-->|journal=J. Hered. |volume=93 |issue=4 |pages=279–82 |year=2002 |pmid=12407215 |url=http://jhered.oxfordjournals.org/cgi/content/full/93/4/279}}</ref> || 36 |
|||
| [[Tibetan fox]] || 36 |
|||
|- |
|||
| [[Domestic cat]]<ref>{{cite journal |author=Nie W, Wang J, O'Brien PC, ''et al'' |title=The genome phylogeny of domestic cat, red panda and five mustelid species revealed by comparative chromosome painting and G-banding |journal=Chromosome Res. |volume=10 |issue=3 |pages=209–22 |year=2002 |pmid=12067210 |doi=10.1023/A:1015292005631}}</ref> || 38 |
|||
| [[Domestic pig]] || 38 |
|||
|- |
|||
| [[Lab mouse]] || 40 |
|||
| [[Lab rat]] || 42 |
|||
|- |
|||
| [[Rabbit]]{{Fact|date=February 2007}} || 44<!-- taxon? --> |
|||
| [[Syrian hamster]] || 44 |
|||
|- |
|||
| [[Hare]]{{Fact|date=February 2007}} || 46<!-- taxon? --> |
|||
| [[Human]]<ref name=Grouchy/> ||46 |
|||
|- |
|||
| [[Gorilla]]s<!--both species-->, [[Chimpanzee]]s<!--both species--><ref name=Grouchy>{{cite journal |author=De Grouchy J |title=Chromosome phylogenies of man, great apes, and Old World monkeys |journal=Genetica |volume=73 |issue=1-2 |pages=37–52 |year=1987 |pmid=3333352}}</ref> || 48 |
|||
| [[Domestic sheep]] || 54 |
|||
|- |
|||
| [[Elephant]]s<ref>{{cite journal |author=Houck ML, Kumamoto AT, Gallagher DS, Benirschke K |title=Comparative cytogenetics of the African elephant (Loxodonta africana) and Asiatic elephant (Elephas maximus) |journal=Cytogenet. Cell Genet. |volume=93 |issue=3-4 |pages=249–52 |year=2001 |pmid=11528120 |doi=10.1159/000056992}}</ref> || 56<!-- taxon? --> |
|||
| [[Cattle|Cow]] || 60 |
|||
|- |
|||
| [[Donkey]] || 62 |
|||
| [[Horse]] || 64 |
|||
|- |
|||
| [[Dog]]<ref>{{cite journal |author=Wayne RK, Ostrander EA |title=Origin, genetic diversity, and genome structure of the domestic dog |journal=Bioessays |volume=21 |issue=3 |pages=247–57 |year=1999 |pmid=10333734 | doi = 10.1002/(SICI)1521-1878(199903)21:3 <!--Retrieved from PMID by DOI bot-->}}</ref> || 78 |
|||
| [[Kingfisher]]<ref>{{cite journal |author=Burt DW |title=Origin and evolution of avian microchromosomes |journal=Cytogenet. Genome Res. |volume=96 |issue=1-4 |pages=97–112 |year=2002 |pmid=12438785 |doi=10.1159/000063018}}</ref> || 132 |
|||
|- |
|||
| [[Goldfish]]<ref>{{cite journal |author=Ciudad J, Cid E, Velasco A, Lara JM, Aijón J, Orfao A |title=Flow cytometry measurement of the DNA contents of G0/G1 diploid cells from three different teleost fish species |journal=Cytometry |volume=48 |issue=1 |pages=20–5 |year=2002 |pmid=12116377 | doi = 10.1002/cyto.10100 <!--Retrieved from PMID by DOI bot-->}}</ref> || 100-104 |
|||
| [[Bombyx mori|Silkworm]]<ref>{{cite journal |author=Yasukochi Y, Ashakumary LA, Baba K, Yoshido A, Sahara K |title=A second-generation integrated map of the silkworm reveals synteny and conserved gene order between lepidopteran insects |journal=Genetics |volume=173 |issue=3 |pages=1319–28 |year=2006 |pmid=16547103 |doi=10.1534/genetics.106.055541}}</ref> || 56 |
|||
|} |
|||
|- |
|||
| colspan="2" | |
|||
{| class="wikitable" style="float:right; margin:1em 0 1em 1em" |
|||
|+ Chromosome numbers in other organisms |
|||
|- |
|||
! Species !! Large<br />Chromosomes !! Intermediate<br />Chromosomes !! Small<br />Chromosomes |
|||
|- |
|||
| ''[[Trypanosoma brucei]]'' || 11 || 6 || ~100 |
|||
|- |
|||
| [[Chicken]]<ref>{{cite journal |author=Smith J, Burt DW |title=Parameters of the chicken genome (Gallus gallus) |journal=Anim. Genet. |volume=29 |issue=4 |pages=290–4 |year=1998 |pmid=9745667 |doi=10.1046/j.1365-2052.1998.00334.x}}</ref> || 8 || 2 sex chromosomes || 60 |
|||
|} |
|||
Normal members of a particular eukaryotic [[species]] all have the same number of nuclear chromosomes (see the table). Other eukaryotic chromosomes, i.e., mitochondrial and plasmid-like small chromosomes, are much more variable in number, and there may be thousands of copies per cell. |
|||
Yesterday there was a new XC-Skiing user box listed in "Snow Sports". As of this morning, it's gone. |
|||
[[Image:PLoSBiol3.5.Fig1bNucleus46Chromosomes.jpg|thumb|200px|left|The 24 human chromosome territories during prometaphase in [[fibroblast]] cells.]] |
|||
I put it on my page yesterday, but I doubt it lasted more than 30 hours or so. |
|||
[[Asexual reproduction|Asexually reproducing]] species have one set of chromosomes, which is the same in all body cells. |
|||
Anyone know what happened to the new box? Can it be restored? |
|||
[[sexual reproduction|Sexually reproducing]] species have [[somatic cell]]s (body cells), which are [[ploidy#diploid|diploid]] [2n] having two sets of chromosomes, one from the mother and one from the father. [[Gamete]]s, reproductive cells, are [[ploidy#haploid and monoploidy|haploid]] [n]: They have one set of chromosomes. Gametes are produced by [[meiosis]] of a diploid [[germ line]] cell. During meiosis, the matching chromosomes of father and mother can exchange small parts of themselves ([[Chromosomal crossover|crossover]]), and thus create new chromosomes that are not inherited solely from either parent. When a male and a female gamete merge ([[fertilization]]), a new diploid organism is formed. |
|||
By the way, XC-Skiing is different from regular ("downhill") skiing, so there was no redundancy. [[User:Joel Russ|Joel Russ]] 19:01, 15 August 2006 (UTC) |
|||
Some animal and plant species are [[ploidy#polyploid|polyploid]] [Xn]: They have more than two sets of [[homologous chromosome]]s. Plants important in agriculture such as [[tobacco]] or [[wheat]] are often polyploid, compared to their ancestral species. Wheat has a haploid number of seven chromosomes, still seen in some [[cultivar]]s as well as the wild progenitors. The more-common [[pasta]] and [[bread]] wheats are polyploid, having 28 (tetraploid) and 42 (hexaploid) chromosomes, compared to the 14 (diploid) chromosomes in the wild wheat.<ref>Sakamura, T. (1918), ''Kurze Mitteilung uber die Chromosomenzahlen und die Verwandtschaftsverhaltnisse der Triticum-Arten''. Bot. Mag., 32: 151-154.</ref> |
|||
== bicyclist userbox glitch? == |
|||
What's up with {{[[User:UBX/Bicyclist]]}} suddenly generating my username at the right-hand margin? |
|||
[[User:DanB DanD|DanB DanD]] 20:33, 15 August 2006 (UTC) |
|||
===Prokaryotes=== |
|||
== How about the UFC? == |
|||
[[Prokaryote]] [[species]] generally have one copy of each major chromosome, but most cells can easily survive with multiple copies.<ref>Charlebois R.L. (ed) 1999. ''Organization of the prokaryote genome''. ASM Press, Washington DC.</ref> For example, ''[[Buchnera]]'', a [[symbiosis|symbiont]] of [[aphid]]s has multiple copies of its chromosome, ranging from 10–400 copies per cell.<ref>{{cite journal |author=Komaki K, Ishikawa H |title=Genomic copy number of intracellular bacterial symbionts of aphids varies in response to developmental stage and morph of their host |journal=Insect Biochem. Mol. Biol. |volume=30 |issue=3 |pages=253–8 |year=2000 |month=March |pmid=10732993 |doi=10.1016/S0965-1748(99)00125-3}}</ref> However, in some large bacteria, such as ''[[Epulopiscium fishelsoni]]'' up to 100,000 copies of the chromosome can be present.<ref>{{cite journal |author=Mendell JE, Clements KD, Choat JH, Angert ER |title=Extreme polyploidy in a large bacterium |journal=Proc. Natl. Acad. Sci. U.S.A. |volume=105 |issue=18 |pages=6730–4 |year=2008 |month=May |pmid=18445653 |doi=10.1073/pnas.0707522105 |url=http://www.pnas.org/cgi/pmidlookup?view=long&pmid=18445653}}</ref> Plasmids and plasmid-like small chromosomes are, as in eukaryotes, very variable in copy number. The number of plasmids in the cell is almost entirely determined by the rate of division of the plasmid - fast division causes high copy number, and vice versa. |
|||
==Karyotype== |
|||
A UFC userbox anyone? [[User:Thebigaster|Thebigaster]] 01:05, 18 August 2006 (UTC) |
|||
{{main|Karyotype}} |
|||
[[Image:NHGRI human male karyotype.png|thumb|right|200px|'''Figure 3''': Karyogram of a human male]] |
|||
In general, the '''karyotype''' is the characteristic chromosome complement of a [[eukaryote]] [[species]].<ref>White M.J.D. 1973. ''The chromosomes''. 6th ed, Chapman & Hall, London. p28</ref> The preparation and study of karyotypes is part of [[cytogenetics]]. |
|||
Although the [[replication]] and [[transcription]] of [[DNA]] is highly standardized in [[eukaryotes]], ''the same cannot be said for their karotypes'', which are often highly variable. There may be variation between species in chromosome number and in detailed organization. In some cases, there is significant variation within species. Often there is 1. variation between the two sexes; 2. variation between the [[germ-line]] and [[soma]] (between [[gametes]] and the rest of the body); 3. variation between members of a population, due to [[polymorphism (biology)|balanced genetic polymorphism]]; 4. [[allopatric speciation|geographical variation]] between [[Race (classification of human beings)|races]]; 5. [[mosaic (genetics)|mosaics]] or otherwise abnormal individuals. Also, variation in karyotype may occur during development from the fertilised egg. |
|||
==Scuba Diving== |
|||
Im padi proffessional but the link is broken. Ne1 mind fixing it? |
|||
The technique of determining the karyotype is usually called ''karyotyping''. Cells can be locked part-way through division (in metaphase) [[in vitro]] (in a reaction vial) with [[colchicine]]. These cells are then stained, photographed, and arranged into a ''karyogram'', with the set of chromosomes arranged, autosomes in order of length, and sex chromosomes (here XY) at the end: Fig. 3. |
|||
== How about bocce? Can someone make a User Box? == |
|||
Like many sexually reproducing species, humans have special [[XY sex-determination system|gonosomes]] (sex chromosomes, in contrast to [[autosome]]s). These are XX in females and XY in males. <!--- Irrelevant in this section:"In females, one of the two X chromosomes is inactive and can be seen under a microscope as [[Barr body|Barr bodies]]."---> |
|||
I just played another game of [[bocce]] a few days ago. Much fun. I got exposed to it as a kid. And quite similar to a French precision ball game I've played locally, too. Would someone make a User Box, please. [[User:Joel Russ|Joel Russ]] 13:52, 27 August 2006 (UTC) |
|||
===Historical note=== |
|||
== Kaka fans userboxes/ How about boxes for players? == |
|||
Investigation into the human karyotype took many years to settle the most basic question. How many chromosomes does a normal [[diploid]] human cell contain? In 1912, [[Hans von Winiwarter]] reported 47 chromosomes in [[spermatogonia]] and 48 in [[oogonia]], concluding an [[XO sex-determination system|XX/XO]] [[sex determination]] mechanism.<ref>von Winiwarter H. 1912. Études sur la spermatogenese humaine. ''Arch. biologie'' '''27''', 93, 147-9.</ref> [[Theophilus Painter|Painter]] in 1922 was not certain whether the diploid number of man is 46 or 48, at first favouring 46.<ref>Painter T.S. 1922. The spermatogenesis of man. ''Anat. Res.'' '''23''', 129.</ref> He revised his opinion later from 46 to 48, and he correctly insisted on man's having an [[XY sex-determination system|XX/XY]] system.<ref>Painter T.S. 1923. Studies in mammalian spermatogenesis II. The spermatogenesis of man. ''J. Exp. Zoology'' '''37''', 291-336.</ref> |
|||
New techniques were needed to definitively solve the problem: |
|||
{{KakaFans}} |
|||
:1. Using cells in culture |
|||
Just insert {{KakaFans]}} (delete the ] between Fans and }}) |
|||
:2. Pretreating cells in a [[Tonicity#Hypotonicity|hypotonic solution]], which swells them and spreads the chromosomes |
|||
P.S.Excuse me for my stupid, how about player userboxes like these? Where can we put them in? |
|||
:3. Arresting [[mitosis]] in [[metaphase]] by a solution of [[colchicine]] |
|||
:4. Squashing the preparation on the slide forcing the chromosomes into a single plane |
|||
:5. Cutting up a photomicrograph and arranging the result into an indisputable karyogram. |
|||
It took until the mid-1950s until it became generally accepted that the human karyotype include only 46 chromosomes. Considering the techniques of Winiwarter and Painter, their results were quite remarkable.<ref>Tjio J.H & Levan A. 1956. The chromosome number of man. ''Hereditas'' '''42''', 1-6.</ref><ref>Hsu T.C. ''Human and mammalian cytogenetics: a historical perspective''. Springer-Verlag, N.Y. p10: "It's amazing that he [Painter] even came close!"</ref> Chimpanzees (the closest living relatives to modern humans) have 48 chromosomes. |
|||
== Curling userbox == |
|||
==Chromosomal aberrations== |
|||
How come there's no curling userbox? That game is just awesome! [[User:Vivi Sparrow|Vivi Sparrow]] 20:29, 4 April 2007 (UTC) |
|||
{{main|Chromosome abnormalities|aneuploidy}} |
|||
[[Image:Single Chromosome Mutations.png|thumb|right|The three major single chromosome mutations; deletion (1), duplication (2) and inversion (3).]] |
|||
[[Image:Two Chromosome Mutations.png|thumb|right|The two major two-chromosome mutations; insertion (1) and translocation (2).]] |
|||
[[Image:Chromosome 21.gif|In Down syndrome, there are three copies of chromosome 21|thumb]] |
|||
Chromosomal aberrations are disruptions in the normal chromosomal content of a cell, and are a major cause of genetic conditions in humans, such as [[Down syndrome]]. Some chromosome abnormalities do not cause disease in carriers, such as [[translocations]], or [[chromosomal inversions]], although they may lead to a higher chance of birthing a child with a chromosome disorder. Abnormal numbers of chromosomes or chromosome sets, [[aneuploidy]], may be lethal or give rise to genetic disorders. [[Genetic counseling]] is offered for families that may carry a chromosome rearrangement. |
|||
== Hockey == |
|||
The box for hockey automatically refers to ice hockey (very US-orientated) and also there is no box for hockey (field hockey to the yanks). [[User:SheffGruff|SheffGruff]] 23:55, 11 October 2007 (UTC) |
|||
The gain or loss of chromosome material can lead to a variety of [[genetic disorders]]. Human examples include: |
|||
* [[Cri du chat]], which is caused by the [[Genetic deletion|deletion]] of part of the short arm of chromosome 5. "Cri du chat" means "cry of the cat" in French, and the condition was so-named because affected babies make high-pitched cries that sound like those of a cat. Affected individuals have wide-set eyes, a small head and jaw, and are moderately to severely mentally retarded and very short. |
|||
* [[Wolf-Hirschhorn syndrome]], which is caused by partial deletion of the short arm of chromosome 4. It is characterized by severe growth retardation and severe to profound mental retardation. |
|||
* [[Down's syndrome]], usually is caused by an extra copy of chromosome 21 ([[trisomy 21]]). Characteristics include decreased muscle tone, stockier build, asymmetrical skull, slanting eyes and mild to moderate mental retardation.<ref>{{cite book|last=Miller|first=Kenneth R.|title=Biology|publisher=Prentice Hall|Location=Upper Saddle River, New Jersey|date=2000|edition=5|pages=194-195|chapter=9-3|isbn=0-13-436265-9|accessdate=2008-03-31|=Add citation}}</ref> |
|||
* [[Edwards syndrome]], which is the second-most-common trisomy; Down syndrome is the most common. It is a trisomy of chromosome 18. Symptoms include mental and motor retardation and numerous congenital anomalies causing serious health problems. Ninety percent die in infancy; however, those that live past their first birthday usually are quite healthy thereafter. They have a characteristic clenched hands and overlapping fingers. |
|||
* [[Patau Syndrome]], also called D-Syndrome or trisomy-13. Symptoms are somewhat similar to those of trisomy-18, but they do not have the characteristic hand shape. |
|||
* [[Idic15]], abbreviation for Isodicentric 15 on chromosome 15; also called the following names due to various researches, but they all mean the same; IDIC(15), Inverted dupliction 15, extra Marker, Inv dup 15, partial tetrasomy 15 |
|||
* [[Jacobsen syndrome]], also called the terminal 11q deletion disorder.<ref>[http://www.11q.org European Chromosome 11 Network<!-- Bot generated title -->]</ref> This is a very rare disorder. Those affected have normal intelligence or mild mental retardation, with poor expressive language skills. Most have a bleeding disorder called [[Paris-Trousseau syndrome]]. |
|||
* [[Klinefelter's syndrome]] (XXY). Men with Klinefelter syndrome are usually sterile, and tend to have longer arms and legs and to be taller than their peers. Boys with the syndrome are often shy and quiet, and have a higher incidence of speech delay and [[dyslexia]]. During puberty, without testosterone treatment, some of them may develop [[gynecomastia]]. |
|||
* [[Turner syndrome]] (X instead of XX or XY). In Turner syndrome, female sexual characteristics are present but underdeveloped. People with Turner syndrome often have a short stature, low hairline, abnormal eye features and bone development and a "caved-in" appearance to the chest. |
|||
* [[XYY syndrome]]. XYY boys are usually taller than their siblings. Like XXY boys and XXX girls, they are somewhat more likely to have learning difficulties. |
|||
* [[Triple-X syndrome]] (XXX). XXX girls tend to be tall and thin. They have a higher incidence of dyslexia. |
|||
* [[Small supernumerary marker chromosome]]. This means there is an extra, abnormal chromosome. Features depend on the origin of the extra genetic material. [[Cat-eye syndrome]] and [[Isodicentric 15|isodicentric chromosome 15 syndrome]] (or Idic15) are both caused by a supernumerary marker chromosome, as is [[Pallister-Killian syndrome]]. |
|||
Chromosomal mutations produce changes in whole chromosomes (more than one gene) or in the number of chromosomes present. |
|||
== Netball Userboxes == |
|||
* Deletion - loss of part of a chromosome |
|||
* Duplication - extra copies of a part of a chromosome |
|||
* Inversion - reverse the direction of a part of a chromosome |
|||
* Translocation - part of a chromosome breaks off and attaches to another chromosome |
|||
Most mutations are neutral - have little or no effect |
|||
A detailed graphical display of all human chromosomes and the diseases annotated at the correct spot may be found at<ref>[http://www.ornl.gov/hgmis/posters/chromosome/ Exploring Genes & Genetic Disorders<!-- Bot generated title -->]</ref>. |
|||
Can someone please create some Netball userboxes. |
|||
[[User:Glamgirljaspreet101|Glamgirljaspreet101]] ([[User talk:Glamgirljaspreet101|talk]]) 00:23, 27 December 2007 (UTC) |
|||
==Human chromosomes== |
|||
:Make your own! They are easy to create - easy to follow instructions are available at [[Wikipedia:Userboxes#Designing_a_userbox]] - [[User:Ahunt|Ahunt]] ([[User talk:Ahunt|talk]]) 00:35, 27 December 2007 (UTC) |
|||
Human cells have 23 pairs of large linear nuclear chromosomes, giving a total of 46 per cell. In addition to these, human cells have many hundreds of copies of the [[mitochondrial genome]]. [[DNA sequencing|Sequencing]] of the human genome has provided a great deal of information about each of the chromosomes. Below is a table compiling statistics for the chromosomes, based on the [[Sanger Institute]]'s human genome information in the Vertebrate Genome Annotation (VEGA) database.<ref>http://vega.sanger.ac.uk/Homo_sapiens/index.html All data in this table was derived from this database, July 7 2007.</ref> Number of genes is an estimate as it is in part based on [[gene prediction]]s. Total chromosome length is an estimate as well, based on the estimated size of unsequenced [[heterochromatin]] regions. |
|||
==Can you create a user box?== |
|||
{| class="wikitable sortable" style="text-align:right" |
|||
If so, how? [[User:Oohoui1123|Oohoui1123]] ([[User talk:Oohoui1123|talk]]) 22:11, 17 August 2008 (UTC) |
|||
|- |
|||
! Chromosome !! [[Genes]] !! Total [[Nucleobase|bases]] !! Sequenced bases<ref>Sequenced percentages are based on fraction of euchromatin portion, as the [[Human Genome Project]] goals called for determination of only the [[euchromatin|euchromatic]] portion of the genome. [[Telomere]]s, [[centromere]]s, and other [[heterochromatin|heterochromatic]] regions have been left undetermined, as have a small number of unclonable gaps. See http://www.ncbi.nlm.nih.gov/genome/seq/ for more information on the Human Genome Project.</ref> |
|||
|- |
|||
| [[Chromosome 1 (human)|1]] || 3,148 || 247,200,000 || 224,999,719 |
|||
|- |
|||
| [[Chromosome 2 (human)|2]] || 902 || 242,750,000 || 237,712,649 |
|||
|- |
|||
| [[Chromosome 3 (human)|3]] || 1,436 || 199,450,000 || 194,704,827 |
|||
|- |
|||
| [[Chromosome 4 (human)|4]] || 453 || 191,260,000 || 187,297,063 |
|||
|- |
|||
| [[Chromosome 5 (human)|5]] || 609 || 180,840,000 || 177,702,766 |
|||
|- |
|||
| [[Chromosome 6 (human)|6]] || 1,585 || 170,900,000 || 167,273,992 |
|||
|- |
|||
| [[Chromosome 7 (human)|7]] || 1,824 || 158,820,000 || 154,952,424 |
|||
|- |
|||
| [[Chromosome 8 (human)|8]] || 781 || 146,270,000 || 142,612,826 |
|||
|- |
|||
| [[Chromosome 9 (human)|9]] || 1,229 || 140,440,000 || 120,312,298 |
|||
|- |
|||
| [[Chromosome 10 (human)|10]] || 1,312 || 135,370,000 || 131,624,737 |
|||
|- |
|||
| [[Chromosome 11 (human)|11]] || 405 || 134,450,000 || 131,130,853 |
|||
|- |
|||
| [[Chromosome 12 (human)|12]] || 1,330 || 132,290,000 || 130,303,534 |
|||
|- |
|||
| [[Chromosome 13 (human)|13]] || 623 || 114,130,000 || 95,559,980 |
|||
|- |
|||
| [[Chromosome 14 (human)|14]] || 886 || 106,360,000 || 88,290,585 |
|||
|- |
|||
| [[Chromosome 15 (human)|15]] || 676 || 100,340,000 || 81,341,915 |
|||
|- |
|||
| [[Chromosome 16 (human)|16]] || 898 || 88,820,000 || 78,884,754 |
|||
|- |
|||
| [[Chromosome 17 (human)|17]] || 1,367 || 78,650,000 || 77,800,220 |
|||
|- |
|||
| [[Chromosome 18 (human)|18]] || 365 || 76,120,000 || 74,656,155 |
|||
|- |
|||
| [[Chromosome 19 (human)|19]] || 1,553 || 63,810,000 || 55,785,651 |
|||
|- |
|||
| [[Chromosome 20 (human)|20]] || 816 || 62,440,000 || 59,505,254 |
|||
|- |
|||
| [[Chromosome 21 (human)|21]] || 446 || 46,940,000 || 34,171,998 |
|||
|- |
|||
| [[Chromosome 22 (human)|22]] || 595 || 49,530,000 || 34,893,953 |
|||
|- |
|||
| [[X chromosome|X (sex chromosome)]] || 1,093 || 154,910,000 || 151,058,754 |
|||
|- |
|||
| [[Y chromosome|Y (sex chromosome)]] || 125 || 57,740,000 || 22,429,293 |
|||
|} |
|||
== |
==See also== |
||
* [[Locus (genetics)|Locus]] (explains gene location nomenclature) |
|||
* [[Sex-determination system]] |
|||
** [[XY sex-determination system]] |
|||
*** [[X chromosome]] |
|||
**** [[X-inactivation]] |
|||
*** [[Y chromosome]] |
|||
**** [[Y-chromosomal Adam]] |
|||
**** [[Y-chromosomal Aaron]] |
|||
* [[Genetic genealogy]] |
|||
** [[Genealogical DNA test]] |
|||
* [[Deletion (genetics)|Genetic deletion]] |
|||
* [[List of number of chromosomes of various organisms]] |
|||
==External links== |
|||
Do we have userboxes for fantasy sports practicioners? I'd create one if there aren't. –'''[[User:Howard the Duck|<font color="#FFA500">Howard</font>]] [[Special:Contributions/Howard the Duck|<font color="#FFA500">the</font>]] [[User talk:Howard the Duck|<font color="#FFA500">Duck</font>]]''' 07:07, 23 September 2008 (UTC) |
|||
* [http://atlasgeneticsoncology.org/Educ/PolyMecaEng.html Chromosome Abnormalities at AtlasGeneticsOncology] |
|||
* [http://gslc.genetics.utah.edu/units/disorders/karyotype/ What Can Our Chromosomes Tell Us?], from the University of Utah's Genetic Science Learning Center |
|||
* [http://gslc.genetics.utah.edu/units/disorders/karyotype/karyotype.cfm Try making a karyotype yourself], from the University of Utah's Genetic Science Learning Center |
|||
* [http://users.rcn.com/jkimball.ma.ultranet/BiologyPages/C/Chromosomes.html Kimballs Chromosome pages] |
|||
* [http://www.genomenewsnetwork.org/categories/index/genome/chromosomes.php Chromosome News from Genome News Network] |
|||
* [http://www.chromosomehelpstation.com/eurochromnet.htm Eurochromnet], European network for Rare Chromosome Disorders on the Internet |
|||
* http://www.ensembl.org [[Ensembl]] project, presenting chromosomes, their [[gene]]s and [[synteny|syntenic]] loci graphically via the web |
|||
* [https://www3.nationalgeographic.com/genographic/index.html Genographic Project] |
|||
* [http://ghr.nlm.nih.gov/ghr/chromosomes Home reference on Chromosomes] from the U.S. National Library of Medicine |
|||
==References== |
|||
:I have never seen any, but then perhaps I am not completely sure what you are referring to? Can you give an example or two? - [[User:Ahunt|Ahunt]] ([[User talk:Ahunt|talk]]) 11:03, 23 September 2008 (UTC) |
|||
{{reflist|2}} |
|||
{{chromo}} |
|||
[[Category:Chromosomes| ]] |
|||
[[Category:Nuclear substructures]] |
|||
[[Category:Cytogenetics]] |
|||
{{Link FA|de}} |
|||
[[ar:صبغي]] |
|||
[[ast:Cromosoma]] |
|||
[[bn:ক্রোমোজোম]] |
|||
[[zh-min-nan:Liám-sek-thé]] |
|||
[[bs:Hromozom]] |
|||
[[bg:Хромозома]] |
|||
[[ca:Cromosoma]] |
|||
[[cs:Chromozóm]] |
|||
[[cy:Cromosom]] |
|||
[[da:Kromosom]] |
|||
[[de:Chromosom]] |
|||
[[et:Kromosoom]] |
|||
[[es:Cromosoma]] |
|||
[[eo:Kromosomo]] |
|||
[[fa:رنگینتن]] |
|||
[[fr:Chromosome]] |
|||
[[gl:Cromosoma]] |
|||
[[ko:염색체]] |
|||
[[hi:गुण सूत्र]] |
|||
[[hr:Kromosomi]] |
|||
[[id:Kromosom]] |
|||
[[it:Cromosoma]] |
|||
[[he:כרומוזום]] |
|||
[[ka:ქრომოსომა]] |
|||
[[ku:Kromozom]] |
|||
[[lv:Hromosoma]] |
|||
[[lt:Chromosoma]] |
|||
[[lmo:Cromusoma]] |
|||
[[hu:Kromoszóma]] |
|||
[[mk:Хромозом]] |
|||
[[ms:Kromosom]] |
|||
[[mn:Хромосом]] |
|||
[[nl:Chromosoom]] |
|||
[[ja:染色体]] |
|||
[[no:Kromosom]] |
|||
[[oc:Cromosòma]] |
|||
[[pl:Chromosom]] |
|||
[[pt:Cromossomo]] |
|||
[[ro:Cromozom]] |
|||
[[ru:Хромосома]] |
|||
[[simple:Chromosome]] |
|||
[[sk:Chromozóm]] |
|||
[[sl:Kromosom]] |
|||
[[sr:Хромозом]] |
|||
[[su:Kromosom]] |
|||
[[fi:Kromosomi]] |
|||
[[sv:Kromosom]] |
|||
[[te:వారసవాహిక]] |
|||
[[th:โครโมโซม]] |
|||
[[vi:Nhiễm sắc thể]] |
|||
[[tr:Kromozom]] |
|||
[[uk:Хромосома]] |
|||
[[ur:لونجسیمہ]] |
|||
[[yi:כראמאזאם]] |
|||
[[zh:染色體]] |
|||
Revision as of 02:23, 10 October 2008
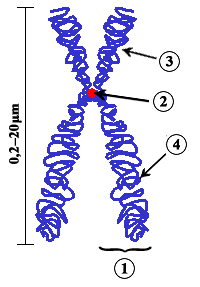
Chromosomes are organized structures of DNA and proteins that are found in cells. A chromosome is a singular piece of DNA, which contains many genes, regulatory elements and other nucleotide sequences. Chromosomes also contain DNA-bound proteins, which serve to package the DNA and control its functions. The word chromosome comes from the Greek χρῶμα (chroma, color) and σῶμα (soma, body) due to their property of being stained very strongly by some dyes.
Chromosomes vary extensively between different organisms. The DNA molecule may be circular or linear, and can contain anything from tens of kilobase pairs to hundreds of megabase pairs. Typically eukaryotic cells (cells with nuclei) have large linear chromosomes and prokaryotic cells (cells without defined nuclei) have smaller circular chromosomes, although there are many exceptions to this rule. Furthermore, cells may contain more than one type of chromosome; for example, mitochondria in most eukaryotes and chloroplasts in plants have their own small chromosomes.
In eukaryotes, nuclear chromosomes are packaged by proteins into a condensed structure called chromatin. This allows the very long DNA molecules to fit into the cell nucleus. The structure of chromosomes and chromatin varies through the cell cycle. Chromosomes may exist as either duplicated or unduplicated—unduplicated chromosomes are single linear strands, whereas duplicated chromosomes (copied during synthesis phase) contain two copies joined by a centromere. Compaction of the duplicated chromosomes during mitosis and meiosis results in the classic four-arm structure (pictured to the right).
"Chromosome" is a rather loosely defined term. In prokaryotes, a small circular DNA molecule may be called either a plasmid or a small chromosome. These small circular genomes are also found in mitochondria and chloroplasts, reflecting their bacterial origins. The simplest chromosomes are found in viruses: these DNA or RNA molecules are short linear or circular chromosomes that often lack any structural proteins.
History
This is a brief history of research in a complex field where each advance was hard won, and often hotly disputed at the time.
Visual discovery of chromosomes. Textbooks have often said that chromosomes were first observed in plant cells by a Swiss botanist named Karl Wilhelm von Nägeli in 1842.[1] However, this opinion has been challenged, perhaps decisively, by Henry Harris, who has freshly reviewed the primary literature.[2] In his opinion the claim of Nägeli to have seen spore mother cells divide is mistaken, as are some of his interpretations. Harris considers other candidates, especially Wilhelm Hofmeister, whose publications in 1848-9 include plates that definitely show mitotic events.[3][4] Hofmeister was also the choice of Cyril Darlington.
The work of other cytologists such as Walther Flemming, Eduard Strasburger, Otto Bütschli, Oskar Hertwig and Carl Rabl should be acknowledged. The use of basophilic aniline dyes was a new technique for effectively staining the chromatin material in the nucleus. Their behavior in animal (salamander) cells was later described in detail by Walther Flemming, who in 1882 "provided a superb summary of the state of the field."[5][6] The name chromosome was invented in 1888 by Heinrich von Waldeyer. However, van Beneden's monograph of 1883 on the fertilised eggs of the parasitic roundworm Ascaris megalocephala was the outstanding work of this period.[7] His conclusions are classic:
- Thus there is no fusion between the male chromatin and the female chromatin at any stage of division....
- The elements of male origin and those of female origin are never fused together in a cleavage nucleus, and perhaps they remain distinct in all the nuclei derived from them. [tranl: Harris p162]
"It is not easy to identify who first discerned chromosomes during mitosis, but there is no doubt that those who first saw them had no idea of their significance... [but] with the work of Balbiani and van Beneden we move away from... the mechanism of cell division to a precise delineation of chromosomes and what they do during the division of the cell."[8]
Van Beneden's master work was closely followed by that of Carl Rabl, who reached similar conclusions. [9] This more or less concludes the first period, in which chromosomes were visually sighted and the morphological stages of mitosis were described. Coleman also gives a useful review of these discoveries.[10]
Nucleus as the seat of heredity. The origin of this epoch-making idea lies in a few sentences tucked away in Ernst Haeckel's Generelle Morphologie of 1866.[11] The evidence for this insight gradually accumulated until, after twenty or so years, two of the greatest in a line of great German scientists spelled out the concept. August Weismann proposed that the germ line is separate from the soma, and that the cell nucleus is the repository of the hereditary material, which, he proposed, is arranged along the chromosomes in a linear manner. Furthermore, he proposed that at fertilisation a new combination of chromosomes (and their hereditary material) would be formed. This was the explanation for the reduction division of meiosis (first described by van Beneden).
Chromosomes as vectors of heredity. In a series of outstanding experiments, Theodor Boveri gave the definitive demonstration that chromosomes are the vectors of heredity. His two principles were:
- The continuity of chromosomes
- The individuality of chromosomes.
It is the second of these principles that was so original. Boveri was able to test the proposal put forward by Wilhelm Roux, that each chromosome carries a different genetic load, and showed that Roux was right. Upon the rediscovery of Mendel, Boveri was able to point out the connection between the rules of inheritance and the behaviour of the chromosomes. It is interesting to see that Boveri influenced two generations of American cytologists: Edmund Beecher Wilson, Walter Sutton and Theophilus Painter were all influenced by Boveri (Wilson and Painter actually worked with him). In his famous textbook The Cell, Wilson linked Boveri and Sutton together by the Boveri-Sutton theory. Mayr remarks that the theory was hotly contested by some famous geneticists: William Bateson, Wilhelm Johannsen, Richard Goldschmidt and T.H. Morgan, all of a rather dogmatic turn-of-mind. Eventually complete proof came from chromosome maps – in Morgan's own lab.[12]
Chromosomes in eukaryotes
It has been suggested that Eukaryotic chromosome fine structure be merged into this article. (Discuss) Proposed since December 2007. |
Eukaryotes (cells with nuclei such as plants, yeast, and animals) possess multiple large linear chromosomes contained in the cell's nucleus. Each chromosome has one centromere, with one or two arms projecting from the centromere, although, under most circumstances, these arms are not visible as such. In addition, most eukaryotes have a small circular mitochondrial genome, and some eukaryotes may have additional small circular or linear cytoplasmic chromosomes.
In the nuclear chromosomes of eukaryotes, the uncondensed DNA exists in a semi-ordered structure, where it is wrapped around histones (structural proteins), forming a composite material called chromatin.
Chromatin

Chromatin is the complex of DNA and protein found in the eukaryotic nucleus which packages chromosomes. The structure of chromatin varies significantly between different stages of the cell cycle, according to the requirements of the DNA.
Interphase chromatin
During interphase (the period of the cell cycle where the cell is not dividing) two types of chromatin can be distinguished:
- Euchromatin, which consists of DNA that is active, e.g., expressed as protein.
- Heterochromatin, which consists of mostly inactive DNA. It seems to serve structural purposes during the chromosomal stages. Heterochromatin can be further distinguished into two types:
- Constitutive heterochromatin, which is never expressed. It is located around the centromere and usually contains repetitive sequences.
- Facultative heterochromatin, which is sometimes expressed.
Individual chromosomes cannot be distinguished at this stage - they appear in the nucleus as a homogeneous tangled mix of DNA and protein.
Metaphase chromatin and division

In the early stages of mitosis or meiosis (cell division), the chromatin strands become more and more condensed. They cease to function as accessible genetic material (transcription stops) and become a compact transportable form. This compact form makes the individual chromosomes visible, and they form the classic four arm structure, a pair of sister chromatids attached to each other at the centromere. The shorter arms are called p arms (from the French petit, small) and the longer arms are called q arms (q follows p in the Latin alphabet). This is the only natural context in which individual chromosomes are visible with an optical microscope.
During divisions long microtubules attach to the centromere and the two opposite ends of the cell. The microtubules then pull the chromatids apart, so that each daughter cell inherits one set of chromatids. Once the cells have divided, the chromatids are uncoiled and can function again as chromatin. In spite of their appearance, chromosomes are structurally highly condensed, which enables these giant DNA structures to be contained within a cell nucleus (Fig. 2).
The self-assembled microtubules form the spindle, which attaches to chromosomes at specialized structures called kinetochores, one of which is present on each sister chromatid. A special DNA base sequence in the region of the kinetochores provides, along with special proteins, longer-lasting attachment in this region.
Chromosomes in prokaryotes
The prokaryotes - bacteria and archaea - typically have a single circular chromosome, but many variations do exist.[13] Most bacteria have a single circular chromosome that can range in size from only 160,000 base pairs in the endosymbiotic bacteria Candidatus Carsonella ruddii,[14] to 12,200,000 base pairs in the soil-dwelling bacteria Sorangium cellulosum.[15] Spirochaetes of the genus Borrelia are a notable exception to this arrangement, with bacteria such as Borrelia burgdorferi, the cause of Lyme disease, containing a single linear chromosome.[16]
Structure in sequences
Prokaryotes chromosomes have less sequence-based structure than eukaryotes. Bacteria typically have a single point (the origin of replication) from which replication starts, whereas some archaea contain multiple replication origins.[17] The genes in prokaryotes are often organized in operons, and do not contain introns, unlike eukaryotes.
DNA packaging
Prokaryotes do not possess nuclei. Instead, their DNA is organized into a structure called the nucleoid.[18] The nucleoid is a distinct structure and occupies a defined region of the bacterial cell. This structure is, however, dynamic and is maintained and remodeled by the actions of a range of histone-like proteins, which associate with the bacterial chromosome.[19] In archaea, the DNA in chromosomes is even more organized, with the DNA packaged within structures similar to eukaryotic nucleosomes.[20][21]
Bacterial chromosomes tend to be tethered to the plasma membrane of the bacteria. In molecular biology application, this allows for its isolation from plasmid DNA by centrifugation of lysed bacteria and pelleting of the membranes (and the attached DNA).
Prokaryotic chromosomes and plasmids are, like eukaryotic DNA, generally supercoiled. The DNA must first be released into its relaxed state for access for transcription, regulation, and replication.
Number of chromosomes in various organisms
Eukaryotes
These tables give the total number of chromosomes (including sex chromosomes) in a cell nucleus. For example, human cells are diploid and have 22 different types of autosomes, each present as two copies, and two sex chromosomes. This gives 46 chromosomes in total. Other organisms have more than two copies of their chromosomes, such as Bread wheat, which is hexaploid and has six copies of seven different chromosomes - 42 chromosomes in total.
|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Normal members of a particular eukaryotic species all have the same number of nuclear chromosomes (see the table). Other eukaryotic chromosomes, i.e., mitochondrial and plasmid-like small chromosomes, are much more variable in number, and there may be thousands of copies per cell.  Asexually reproducing species have one set of chromosomes, which is the same in all body cells. Sexually reproducing species have somatic cells (body cells), which are diploid [2n] having two sets of chromosomes, one from the mother and one from the father. Gametes, reproductive cells, are haploid [n]: They have one set of chromosomes. Gametes are produced by meiosis of a diploid germ line cell. During meiosis, the matching chromosomes of father and mother can exchange small parts of themselves (crossover), and thus create new chromosomes that are not inherited solely from either parent. When a male and a female gamete merge (fertilization), a new diploid organism is formed. Some animal and plant species are polyploid [Xn]: They have more than two sets of homologous chromosomes. Plants important in agriculture such as tobacco or wheat are often polyploid, compared to their ancestral species. Wheat has a haploid number of seven chromosomes, still seen in some cultivars as well as the wild progenitors. The more-common pasta and bread wheats are polyploid, having 28 (tetraploid) and 42 (hexaploid) chromosomes, compared to the 14 (diploid) chromosomes in the wild wheat.[40] ProkaryotesProkaryote species generally have one copy of each major chromosome, but most cells can easily survive with multiple copies.[41] For example, Buchnera, a symbiont of aphids has multiple copies of its chromosome, ranging from 10–400 copies per cell.[42] However, in some large bacteria, such as Epulopiscium fishelsoni up to 100,000 copies of the chromosome can be present.[43] Plasmids and plasmid-like small chromosomes are, as in eukaryotes, very variable in copy number. The number of plasmids in the cell is almost entirely determined by the rate of division of the plasmid - fast division causes high copy number, and vice versa. Karyotype In general, the karyotype is the characteristic chromosome complement of a eukaryote species.[44] The preparation and study of karyotypes is part of cytogenetics. Although the replication and transcription of DNA is highly standardized in eukaryotes, the same cannot be said for their karotypes, which are often highly variable. There may be variation between species in chromosome number and in detailed organization. In some cases, there is significant variation within species. Often there is 1. variation between the two sexes; 2. variation between the germ-line and soma (between gametes and the rest of the body); 3. variation between members of a population, due to balanced genetic polymorphism; 4. geographical variation between races; 5. mosaics or otherwise abnormal individuals. Also, variation in karyotype may occur during development from the fertilised egg. The technique of determining the karyotype is usually called karyotyping. Cells can be locked part-way through division (in metaphase) in vitro (in a reaction vial) with colchicine. These cells are then stained, photographed, and arranged into a karyogram, with the set of chromosomes arranged, autosomes in order of length, and sex chromosomes (here XY) at the end: Fig. 3. Like many sexually reproducing species, humans have special gonosomes (sex chromosomes, in contrast to autosomes). These are XX in females and XY in males. Historical noteInvestigation into the human karyotype took many years to settle the most basic question. How many chromosomes does a normal diploid human cell contain? In 1912, Hans von Winiwarter reported 47 chromosomes in spermatogonia and 48 in oogonia, concluding an XX/XO sex determination mechanism.[45] Painter in 1922 was not certain whether the diploid number of man is 46 or 48, at first favouring 46.[46] He revised his opinion later from 46 to 48, and he correctly insisted on man's having an XX/XY system.[47] New techniques were needed to definitively solve the problem:
It took until the mid-1950s until it became generally accepted that the human karyotype include only 46 chromosomes. Considering the techniques of Winiwarter and Painter, their results were quite remarkable.[48][49] Chimpanzees (the closest living relatives to modern humans) have 48 chromosomes. Chromosomal aberrations   Chromosomal aberrations are disruptions in the normal chromosomal content of a cell, and are a major cause of genetic conditions in humans, such as Down syndrome. Some chromosome abnormalities do not cause disease in carriers, such as translocations, or chromosomal inversions, although they may lead to a higher chance of birthing a child with a chromosome disorder. Abnormal numbers of chromosomes or chromosome sets, aneuploidy, may be lethal or give rise to genetic disorders. Genetic counseling is offered for families that may carry a chromosome rearrangement. The gain or loss of chromosome material can lead to a variety of genetic disorders. Human examples include:
Chromosomal mutations produce changes in whole chromosomes (more than one gene) or in the number of chromosomes present.
Most mutations are neutral - have little or no effect A detailed graphical display of all human chromosomes and the diseases annotated at the correct spot may be found at[52]. Human chromosomesHuman cells have 23 pairs of large linear nuclear chromosomes, giving a total of 46 per cell. In addition to these, human cells have many hundreds of copies of the mitochondrial genome. Sequencing of the human genome has provided a great deal of information about each of the chromosomes. Below is a table compiling statistics for the chromosomes, based on the Sanger Institute's human genome information in the Vertebrate Genome Annotation (VEGA) database.[53] Number of genes is an estimate as it is in part based on gene predictions. Total chromosome length is an estimate as well, based on the estimated size of unsequenced heterochromatin regions.
See also
External links
References
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
