Cas9
| Cas9 | ||
|---|---|---|

|
||
| CRISPR-associated protein Cas9 from Streptococcus pyogenes | ||
| other names |
SpyCas9, Cas5, Csn1, Csx12 |
|
| Mass / length primary structure | 1,368 amino acids , 158,441 Da | |
| Identifier | ||
| External IDs | ||
| Enzyme classification | ||
| EC, category | 3.1.-.- | |
Cas9 (from English CRISPR-associated , outdated also Cas5, CSN1 or Csx12 ) is an endonuclease and a ribonucleoprotein from bacteria .
properties
The CRISPR / Cas system comes from an adaptive antiviral defense mechanism from bacteria, the CRISPR . The endonuclease Cas9 can bind a certain RNA sequence ( crRNA repeat , sequence GUUUUAGAGCU (A / G) UG (C / U) UGUUUUG) and cut DNA. This crRNA repeat sequence forms an RNA secondary structure , binds a tracrRNA and is then bound by Cas9 . The crRNA repeat sequence is then followed by a sequence that binds to the target DNA ( crRNA spacer ); both sequences are referred to together as crRNA . The second part is the crRNA spacer sequence in the function of a variable adapter, which is complementary to the target DNA and binds to the target DNA. This will cut the DNA near the binding site. Anti-CRISPR proteins are formed by bacteriophages to inhibit Cas proteins .
structure
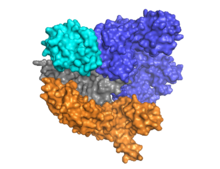
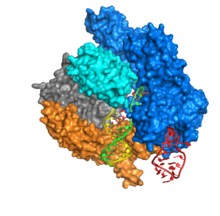
Cas9 consists of two areas, with the RNA embedded between the α-helical area (blue) and the area of the endonuclease (cyan, orange, gray). The two areas are connected by a helix. The nuclease area contains two endonuclease protein domains , RuvC (gray) and the HNH nuclease (cyan). The HNH nuclease cuts the DNA strand bound to the crRNA , while the RuvC cuts the opposite strand. The DNA to be cut must contain the Protospacer adjacent motif (PAM), which consists of the three nucleotides NGG and is bound by the PAM interacting domain at the C terminus of Cas9 (PI domain, orange). Cas9 has three different conformations , unbound, bound to RNA, and bound to RNA and DNA.
Cas9 binds the stem-loop secondary structure of the crRNA. The complex of Cas9, crRNA and tracrRNA binds to the target DNA and cuts both DNA strands. The two RNAs can also be accommodated in a single, self-hybridizing strand in sections ( single-guide RNA , sgRNA). The DNA binding domain ( recognition domain , REC) and the endonuclease domain (NUC) are spatially separated. Since the HNH domain is flexibly connected to the rest of the protein, it cannot be seen in the Cas9 protein crystal . Cas9 uses manganese ions as cofactors .
Types
There are more than 40 different Cas protein families. The families can be divided into three types. Type I, II and IIIa bind and cut double-stranded DNA, while type IIIb binds and cuts single-stranded RNA. In all types, the spacer is formed in bacteria by Cas1 and Cas2 . In types IA and IE, the DNA cut is made by Cas3 , while in type II Cas9 , in type III-A Csm6 and in type III-B Cmr4 causes the cut. Types I and III are structurally related, which suggests a common origin. The helical protein Cas of type III has several β-hairpins that push the double helix of the crRNA and the target RNA apart for the cut at intervals of six nucleotides.
Applications
Cas9 is used as part of the CRISPR / Cas system for genome editing .
Individual evidence
- ^ R. Sorek, V. Kunin, P. Hugenholtz: CRISPR - a widespread system that provides acquired resistance against phages in bacteria and archaea. In: Nature reviews. Microbiology. Volume 6, Number 3, March 2008, pp. 181-186, ISSN 1740-1534 . doi: 10.1038 / nrmicro1793 . PMID 18157154 .
- ↑ D. Rath, L. Amlinger, A. Rath, M. Lundgren: The CRISPR-Cas immune system: biology, mechanisms and applications. In: Biochemistry. Volume 117, October 2015, pp. 119-128, doi: 10.1016 / j.biochi.2015.03.025 , PMID 25868999 (review).
- ↑ G. Gasiunas, R. Barrangou , P. Horvath, V. Siksnys: Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. In: Proceedings of the National Academy of Sciences . Volume 109, Number 39, September 2012, pp. E2579-E2586, ISSN 1091-6490 . doi: 10.1073 / pnas.1208507109 . PMID 22949671 . PMC 3465414 (free full text).
- ^ V. Kunin, R. Sorek, P. Hugenholtz: Evolutionary conservation of sequence and secondary structures in CRISPR repeats. In: Genome biology. Volume 8, Number 4, 2007, p. R61, ISSN 1465-6914 . doi: 10.1186 / gb-2007-8-4-r61 . PMID 17442114 . PMC 1896005 (free full text).
- ↑ Wiedenheft B, Sternberg SH, Doudna JA: RNA-guided genetic silencing systems in bacteria and archaea . In: Nature . 482, No. 7385, Feb 2012, pp. 331-8. bibcode : 2012Natur.482..331W . doi : 10.1038 / nature10886 . PMID 22337052 .
- ^ Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F: Genome engineering using the CRISPR-Cas9 system . In: Nat Protoc . 8, No. 11, 2013, pp. 2281-308. doi : 10.1038 / nprot.2013.143 . PMID 24157548 . PMC 3969860 (free full text).
- ↑ Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E: A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity . In: Science . 337, No. 6096, 2012, pp. 816-21. bibcode : 2012Sci ... 337..816J . doi : 10.1126 / science.1225829 . PMID 22745249 .
- ↑ M. Jinek, F. Jiang, DW Taylor, SH Sternberg, E. Kaya, E. Ma, C. Anders, M. Hauer, K. Zhou, S. Lin, M. Kaplan, AT Iavarone, E. Charpentier, E. Nogales, JA Doudna: Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. In: Science . Volume 343, number 6176, March 2014, p. 1247997, doi : 10.1126 / science.1247997 , PMID 24505130 , PMC 4184034 (free full text).
- ↑ DH Haft, J. Selengut, EF Mongodin, KE Nelson: A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR / Cas subtypes exist in prokaryotic genomes. In: PLoS computational biology. Volume 1, number 6, November 2005, p. E60, ISSN 1553-7358 . doi: 10.1371 / journal.pcbi.0010060 . PMID 16292354 . PMC 1282333 (free full text).
- ↑ a b c d D. W. Taylor, Y. Zhu, RHJ Staals, JE Kornfeld, A. Shinkai, J. van der Oost, E. Nogales, JA Doudna: Structures of the CRISPR-Cmr complex reveal mode of RNA target positioning. In: Science. 348, 2015, p. 581, doi: 10.1126 / science.aaa4535 .
- ↑ a b c J. van der Oost, ER Westra, RN Jackson, B. Wiedenheft: Unraveling the structural and mechanistic basis of CRISPR-Cas systems. In: Nature reviews. Microbiology. Volume 12, number 7, July 2014, pp. 479-492, doi: 10.1038 / nrmicro3279 , PMID 24909109 , PMC 4225775 (free full text) (review).