Dam methylase
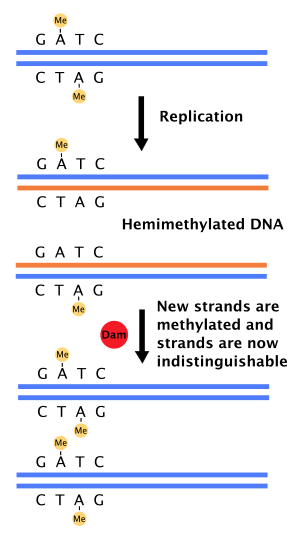
The Dam methylase (abbreviation for deoxyadenosine - methylase ) is an enzyme which has a methyl group to the adenine residue of the GATC sequence in bacterial DNA coupled. The daughter strand is not methylated immediately, which the cell can use for repairs.
Role in the repair of base mismatches
If a base pairing error occurs during DNA synthesis , or if an additional base is incorporated, the molecular cell machinery directs the correction of a DNA mismatch repair protein . For this it is necessary to be able to distinguish the template from the newly synthesized daughter strand. In bacteria , the DNA strands are methylated by Dam methylase. After new synthesis initially only the template has a methyl group; therefore one speaks of the state of "hemimethylation". A repair enzyme binds to mismatched bases and recruits the MutL factor, which then activates the MutH endonuclease . MutH binds hemimethylated DNA specifically to the GATC sequence and cuts the daughter strand here. This enables the unwinding enzyme helicase and exonucleases to remove the faulty sites in the newly synthesized strand. The new synthesis of the removed strand section is carried out by DNA polymerase III.
Role in regulating replication
The hybridization of DNA polymerase with the origin of replication ( oriC ) in bacteria is strictly controlled to ensure that DNA synthesis only takes place once per cell cycle (otherwise the cell would have a multiple of the usual genetic information ). The control takes place, among other things, through the slow hydrolysis of ATP by DnaA . DnaA is a protein that binds to repeats in the oriC to start replication. Dam methylase also plays a role, as oriC has eleven GATC sequences in Escherichia coli . Shortly after the completion of the DNA synthesis, oriC is hemimethylated and is bound by the protein SeqA. The binding of SeqA is specific to hemimethylated GATCs. In this way SeqA blocks the oriC and prevents binding of DnaA. After some time, SeqA separates from the oriC, the GATCs can be re-methylated and bind DnaA again.
Role in the regulation of gene expression
Dam methylase also plays a role in the activation and repression of transcription . In E. coli , GATC sequences are methylated downstream and thus promote transcription. For example, in that it has been described uropathogenic Coli bacteria methylation of two GATC sites next to the PAP - promoter is regulated.
See also
Individual evidence
- ↑ Marinus MG, Morris NR: Isolation of deoxyribonucleic acid methylase mutants of Escherichia coli K-12 . In: J. Bacteriol. . 114, No. 3, June 1973, pp. 1143-50. PMID 4576399 . PMC 285375 (free full text).
- ↑ Geier GE, Modrich P: Recognition sequence of the dam methylase of Escherichia coli K12 and mode of cleavage of Dpn I endonuclease . In: J. Biol. Chem. . 254, No. 4, February 1979, pp. 1408-13. PMID 368070 .
- ↑ a b Barras F, Marinus MG: The great GATC: DNA methylation in E. coli . In: Trends in Genetics . 5, 1989, pp. 139-143. doi : 10.1016 / 0168-9525 (89) 90054-1 .
- ↑ Løbner-Olesen A, Skovgaard O, Marinus, MG: Dam methylation: coordinating cellular processes . In: Curr. Opin. Microbiol. . 8, No. 2, April 2005, pp. 154-160. doi : 10.1016 / j.mib.2005.02.009 . PMID 15802246 .
- ↑ Waldminghaus T and Skarstad K: The Escherichia coli SeqA protein. Plasmid. 2009 May, 61 (3): 141-50. doi : 10.1016 / j.plasmid.2009.02.004
- ↑ Casadesús, J and Low, D: Epigenetic Gene Regulation in the Bacterial World. Microbiol Mol Biol Rev. 2006 Sep; 70 (3): 830-856. doi : 10.1128 / MMBR.00016-06