Foci
| Type-2 restriction enzyme FokI | ||
|---|---|---|
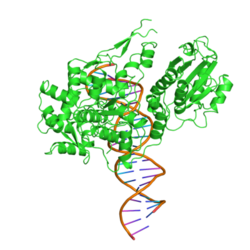
|
||
| FokI bound to DNA, according to PDB 1FOK | ||
| other names |
Endonuclease FokI, Type II restriction enzyme FokI, Type IIS restriction enzyme FokI |
|
| Mass / length primary structure | 583 amino acids , 66,219 Da | |
| Secondary to quaternary structure | Homodimer | |
| Identifier | ||
| External IDs | ||
| Enzyme classification | ||
| EC, category | 3.1.21.4 | |
FokI is a restriction enzyme from the bacterium Flavobacterium okeanokoites .
properties
FokI is an endonuclease (type II, subtype S) which cuts dsDNA according to the recognition sequence 5'-GGATG-3 ', depending on the strand 9 or 13 nucleotides behind the recognition sequence . The cut of the double-stranded DNA by FokI results in a sticky end (end with 4 nucleotides overhang) and a phosphate group each at both 5 'ends of the double-stranded DNA products. FokI is inhibited by DCM methylation and by CpG methylation, but not by DAM methylation . After a restriction of DNA in vitro , FokI can be denatured and thus inactivated by heating it to 65 ° C for 20 minutes . Most of the time, FokI is produced as a recombinant protein in E.coli . FokI cuts dsDNA with two recognition sequences in any orientation better than with one, whereby they can be several hundred nucleotides apart.
Applications
FokI is used for restriction digestions in the context of cloning or restriction analyzes (especially of polymorphisms of the gene of the vitamin D receptor to determine susceptibility to tuberculosis and kidney failure ). The enzymatic part of FokI is used as a fusion protein in the course of a protein design with the recognition domains of other restriction enzymes , e.g. B. for the generation of zinc finger nucleases or in combination with an inactivated Cas9 .
Individual evidence
- ↑ DA Wah, J. Bitinaite, I. Schildkraut, AK Aggarwal: Structure of FokI has implications for DNA cleavage. In: Proceedings of the National Academy of Sciences . Volume 95, Number 18, September 1998, pp. 10564-10569, PMID 9724743 , PMC 27934 (free full text).
- ^ A b S. E. Halford, LE Catto, C. Pernstich, DA Rusling, KL Sanders: The reaction mechanism of FokI excludes the possibility of targeting zinc finger nucleases to unique DNA sites. In: Biochemical Society transactions. Volume 39, Number 2, April 2011, pp. 584-588, doi : 10.1042 / BST0390584 , PMID 21428944 .
- ^ Y. Cao, X. Wang, Z. Cao, X. Cheng: Vitamin D receptor gene FokI polymorphisms and tuberculosis susceptibility: a meta-analysis. In: Archives of Medical Science. Volume 12, number 5, October 2016, pp. 1118–1134, doi : 10.5114 / aoms.2016.60092 , PMID 27695504 , PMC 5016579 (free full text).
- ↑ L. Yang, L. Wu, Y. Fan, J. Ma: Associations among four polymorphisms (BsmI, FokI, TaqI and ApaI) of vitamin D receptor gene and end-stage renal disease: a meta-analysis. In: Archives of medical research. Volume 46, number 1, January 2015, pp. 1-7, doi : 10.1016 / j.arcmed.2014.11.017 , PMID 25434518 .
- ↑ Tsai, SQ et al. (2014). Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nature Biotechnol. 32, 569-576 doi : 10.1038 / nbt.2908
- ↑ Guilinger, JP, Thompson, DB & Liu, DR (2014). Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nature Biotechnol. 32, 577-582 doi : 10.1038 / nbt.2909
- ↑ Wyvekens, N., Topkar, VV, Khayter, C., Joung, JK & Tsai, SQ (2015). Dimeric CRISPR RNA-guided FokI-dCas9 nucleases directed by truncated gRNAs for highly specific genome editing. Hum. Gene Ther. 26, 425-431 doi : 10.1089 / hum.2015.084