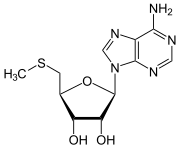
Methionine salvage
| Parent |
| Methionine Synthesis |
| Gene Ontology |
|---|
| QuickGO |
The methionine salvage - pathway (rare: Yang cycle ) is a sequence of six chemical reactions that the starting product 5'-methylthioadenosine (MTA) in the final product L - methionine convert. The enzymes that catalyze the individual reactions can be found in all living things (apart from a few types of bacteria ). The process takes place entirely in the cytosol . At least in the organisms that rely on methionine, it is vital for the recovery of the sulfur atom, the assimilation of which is energy-intensive. The metabolic pathway was first extensively investigated in Klebsiella pneumoniae and Saccharomyces cerevisiae . It was also investigated in plants in connection with ethene biosynthesis. Not all individual steps are fully understood in humans.
MTA is created during the synthesis of polyamines from adenosylmethionine (SAM) or decarboxy-SAM by transferring an aminopropyl group, part of the methionine, of which the methylthio group remains. After the adenine has been split off, this aminopropyl group is then gradually restored at the expense of the ribose ring.
5'-methylthioribose-1-phosphate
In the first reaction, the enzyme 5'-methylthioadenosine phosphorylase ( EC 2.4.2.28 ) catalyzes the cleavage of adenine and simultaneous phosphorylation of MTA to 5'-methylthioribose-1-phosphate. In some bacteria and plants, the enzyme activity is divided between two enzymes, a methylthioadenosine nucleosidase ( EC 3.2.2.16 ) and a methylthioribose kinase ( EC 2.7.1.100 ), the latter, however, consuming one molecule of ATP.
5'-methylthioribulose-1-phosphate
Between 5'-methylthioribose-1-phosphate and 5'-methylthioribulose-1-phosphate there is an equilibrium catalyzed by 5'-methylthioribose-1-phosphate isomerase ( EC 5.3.1.23 ).
2,3-diketo-5'-methylthiopentane-1-phosphate
The dehydration of 5'-methylthioribulose-1-phosphate with the help of 5'-methylthioribulose-1-phosphate dehydratase ( EC 4.2.1.109 ) is difficult to reverse. In humans, this reaction is likely the product of APIP - gene catalyzes the orthologous to the well-studied Mde1p gene of baker's yeast.
Acireductone
The enzyme enolase phosphatase E1 (also: acireductone synthase, EC 3.1.3.77 ) catalyzes the dephosphorylation and subsequent conversion to enol with the result 1,2-dihydroxy-3-keto-5'-methylthiopentene ( acireductone ).
4-methylthio-2-ketobutanoate (MOB)
In a further step, acireductone is oxidized using acireductone dioxygenase and dioxygen, whereby two reaction paths are possible, depending on whether the enzyme carries iron (II) or nickel (II) as a cofactor . With nickel, 3-methylthiopropionate, carbon monoxide and formate ( EC 1.13.11.53 ) are formed - this reaction has been proven in Klebsiella . 4-methylthio-2-ketobutanoate (MOB, KMTB) and formate ( EC 1.13.11.54 ) are formed with iron . It is unclear whether nickel plays a role as a cofactor in humans.
Methionine
 + R-CH (NH 3 + ) -COO - + R-CO-COO -
+ R-CH (NH 3 + ) -COO - + R-CO-COO - ![]()
Finally, MOB is transaminated to methionine. Transaminases can have a wide range of substrates: according to studies in yeast, Aro8p, Aro9p, Bat1p and Bat2p act as MOB transaminases; in the parasite Crithidia fasciculata this is aspartate transaminase, which is orthologous to human gamma-glutamyltransferase . A study on rats showed that several transaminases in the liver cytosol are responsible for methionine synthesis, while mitochondrial transaminases are responsible for the reverse direction, processing excess methionine. Unfortunately, there is no research into the MOB transaminase activity of human transaminases.
regulation
The activity of the metabolic pathway is in principle dependent on the starting substrate MTA. However, an excess of methionine can upset the transamination balance and lead to an excess of MOB, which is likely to be decarboxylated and further broken down in the mitochondria.
Tissue specificity of the methionine salvage metabolic pathway
Originally, it was assumed that the methionine salvage metabolic pathway takes place in all cells of plants. Molecular biological studies in thale cress (Arabidopsis thaliana) and the great plantain (Plantago major) showed, however, that the genes for the enzymes involved in this metabolic pathway are only expressed in the phloem.
Individual evidence
- ^ Wray JW, Abeles RH: The methionine salvage pathway in Klebsiella pneumoniae and rat liver. Identification and characterization of two novel dioxygenases . In: J. Biol. Chem. . 270, No. 7, February 1995, pp. 3147-53. PMID 7852397 .
- ↑ a b c Pirkov I, J Norbeck, Gustafsson L, Albers E: A complete inventory of all enzymes in the eukaryotic methionine salvage pathway . In: FEBS J . . 275, No. 16, August 2008, pp. 4111-20. doi : 10.1111 / j.1742-4658.2008.06552.x . PMID 18625006 .
- ↑ Miyazaki JH, Yang SF: The methionine salvage pathway in relation to ethylene and polyamine biosynthesis . In: Phys. Plant. . 69, No. 2, February 1987, pp. 366-370. doi : 10.1111 / j.1399-3054.1987.tb04302.x .
- ↑ Kamatani N, Nelson-Rees WA, Carson DA: Selective killing of human malignant cell lines deficient in methylthioadenosine phosphorylase, a purine metabolic enzyme . In: Proc. Natl. Acad. Sci. USA . 78, No. 2, February 1981, pp. 1219-23. PMID 6785752 . PMC 319979 (free full text).
- ↑ Kabuyama Y, Litman ES, Templeton PD, et al. : A mediator of Rho-dependent invasion moonlights as a methionine salvage enzyme . In: Mol. Cell Proteomics . 8, No. 10, October 2009, pp. 2308-20. doi : 10.1074 / mcp.M900178-MCP200 . PMID 19620624 . PMC 2758758 (free full text).
- ↑ UniProt Q96GX9
- ↑ Wang H, Pang H, Bartlam M, Rao Z: Crystal structure of human E1 enzyme and its complex with a substrate analog reveals the mechanism of its phosphatase / enolase activity . In: J. Mol. Biol. . 348, No. 4, May 2005, pp. 917-26. doi : 10.1016 / j.jmb.2005.01.072 . PMID 15843022 .
- ↑ Ju T, Goldsmith RB, Chai SC, Maroney MJ, Pochapsky SS, Pochapsky TC: One protein, two enzymes revisited: a structural entropy switch interconverts the two isoforms of acireductone dioxygenase . In: J. Mol. Biol. . 363, No. 4, November 2006, pp. 823-34. doi : 10.1016 / j.jmb.2006.08.060 . PMID 16989860 . PMC 1808343 (free full text).
- ↑ Oram SW, Ai J, Pagani GM, et al. : Expression and function of the human androgen-responsive gene ADI1 in prostate cancer . In: Neoplasia . 9, No. 8, August 2007, pp. 643-51. PMID 17786183 . PMC 1950434 (free full text).
- ↑ Berger LC, Wilson J, Wood P, Berger BJ: Methionine regeneration and aspartate aminotransferase in parasitic protozoa . In: J. Bacteriol. . 183, No. 15, August 2001, pp. 4421-34. doi : 10.1128 / JB.183.15.4421-4434.2001 . PMID 11443076 . PMC 95336 (free full text).
- ↑ a b Scislowski PW, Pickard K: Methionine transamination - metabolic function and Subcellular compartmentation . In: Mol. Cell. Biochem. . 129, No. 1, December 1993, pp. 39-45. PMID 8177225 .
- ↑ Jones SM, Yeaman SJ: Oxidative decarboxylation of 4-methylthio-2-oxobutyrate by branched-chain 2-oxo acid dehydrogenase complex . In: Biochem. J. . 237, No. 2, July 1986, pp. 621-3. PMID 3800905 . PMC 1147032 (free full text).
- ↑ Pommerrenig B, Feussner K, Zierer W, Rabinovych V, Klebl F, Feussner I, Sauer N: Phloem-specific expression of Yang cycle genes and identification of novel Yang cycle enzymes in Plantago and Arabidopsis . In: The Plant Cell . 23, No. 5, May 2011, pp. 1904-19. PMID 21540433 .