NAIL-MS
NAIL-MS (abbreviation for nucleic acid isotope labeling coupled mass spectrometry ) is a technique based on mass spectrometry for the investigation of nucleic acids and their modifications . NAIL-MS enables a variety of experimental designs to study the underlying mechanisms of RNA biology in vivo . For example, the dynamic behavior of nucleic acids, in particular of RNA modifications , can be examined more closely in living cells.
theory
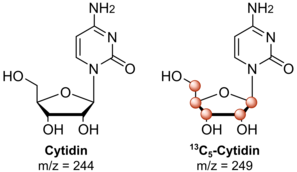
NAIL-MS is used to study RNA modification mechanisms. For this purpose, cells in culture are first fed with stable isotope-labeled nutrients, which the cells incorporate into their biomolecules. After the nucleic acids, mostly RNA , have been purified , analysis is carried out using mass spectrometry. Mass spectrometry is an analytical technique that measures the mass-to-charge ratio of ions . Pairs of chemically identical nucleosides with different compositions of the stable isotopes can be differentiated in a mass spectrometer on the basis of their mass difference. Unlabeled nucleosides can therefore be differentiated from their isotopologues labeled with stable isotopes. For most NAIL-MS approaches it is crucial that the labeled nucleosides are more than 2 Da heavier than the unlabelled ones. This is because 1.1% of naturally occurring carbon atoms are 13 C isotopes. In the case of nucleosides, this leads to a mass increase of 1 Da in ~ 10% of the nucleosides. This signal would be superimposed with the NAIL-MS labeling and thus falsify the evaluation.
NAIL-MS can be used to study the RNA modification dynamics by changing the labeled nutrients of the appropriate growth medium during the experiment. In addition, cell populations can be compared directly with one another without any potential falsification due to individual purification. In addition, NAIL-MS can be used for the production of biosynthetic isotopologues of most nucleosides, which are required for absolute quantification by mass spectrometry and even for the discovery of as yet unknown RNA modifications.
Basic procedure
In general, the cells are cultured in unlabeled or stable (non-radioactive) isotope-labeled media. For example, glucose , which is marked with six carbon-13 atoms ( 13 C) instead of the normal carbon-12 ( 12 C), can be added to the medium . Cells that grow in this medium will, depending on the model organism, incorporate the heavy glucose into all of their RNA molecules. According to this, all nucleotides are 5 Da heavier than their unlabeled isotopologues due to complete carbon labeling of the ribose. After the cells have been cultivated and appropriately labeled, they are generally harvested using phenol / chloroform / guanidinium isothiocyanate extraction . Other extraction methods are possible and sometimes required (e.g. for yeast). The RNA is then isolated via isopropanol or ethanol precipitation. The further purification of specific RNA species (e.g. rRNA or tRNA ) is usually done by size exclusion chromatography (SEC), but other approaches are also possible. For most applications, the end product must be enzymatically digested to nucleosides prior to analysis by LC-MS . Hence, digestive enzymes such as Benzonase, NP1 and CIP are used. A triple quadrupole mass spectrometer in MRM mode is typically used for the measurements.
Labeling the cells
How the labeling of RNA molecules is achieved depends on the model organism. The minimal medium M9 can be used for E. coli (bacteria) and supplemented with the stable isotope-labeled variants of the required salts. This enables labeling with 13 C-carbon, 15 N-nitrogen, 34 S-sulfur and 2 H-hydrogen. With S. cerevisiae (yeast) there are currently two options: On the one hand, the use of commercially available complete medium , which enables labeling with 13 C carbon and / or 15 N nitrogen, and on the other hand, the use of minimal YNB medium , which must be supplemented with several amino acids and glucose, which can be added as stable isotope-labeled variants in order to achieve 13 C-carbon, 15 N-nitrogen and 2 H-hydrogen labeling of the RNA.
While the marking in model organisms such as E. coli and S. cerevisiae is comparatively simple, marking with the aid of stable isotopes in cell culture is a greater challenge, since the composition of the growth media is more complex. Neither the addition of labeled glucose nor the variants of simple precursors of nucleoside biosynthesis such as glutamine and / or aspartate labeled with stable isotopes lead to a defined mass increase of more than 2 Da. While suitable compounds for complete labeling in cell culture have already been found, these results have not yet been published.
Applications
NAIL-MS enables various experiments to be designed.
Manufacture of SILIS
NAIL-MS can be used to produce stable isotope-labeled internal standards (ISTD). For this purpose, cells are grown in medium, which leads to a complete labeling of all nucleosides. The purified nucleoside mixture can then be used as the ISTD, which is necessary for an accurate absolute quantification of the nucleosides by mass spectrometry. This mixture of labeled nucleosides is also known as SILIS (stable isotope labeled internal standard). The advantage of this approach is that all modifications present in an organism can be produced biosynthetically as isotope-labeled compounds. SILIS was manufactured even before the term NAIL-MS came up.
Comparative experiments
A comparative NAIL-MS experiment is very similar to a SILAC experiment, but for RNA instead of proteins. First, two populations of the respective cells are cultivated. One of the cell populations is fed growth medium containing unlabeled nutrients, while the second population is fed growth medium containing stable isotopically labeled nutrients. The cells then incorporate the respective isotopologues into their RNA molecules. One of the cell populations serves as a control group, while the other is subjected to the associated investigations (e.g. KO strain, stress). After the two cell populations have been harvested, they are mixed and processed together in order to exclude falsification through individual purification. Due to the different mass-to-charge ratios of the incorporated nucleosides, a mass spectrometric differentiation between the two cell populations is possible.
Pulse-chase experiments
When initiating a pulse-chase experiment, the medium is changed from medium (1) to medium (2). The two media may only differ in their isotope content. This makes it possible to differentiate between RNA molecules that were already present before the start of the experiment (= RNA molecules grown in the medium (1)) and RNA molecules that are re- transcribed after the start of the experiment (= RNA molecules grown in the medium RNA molecules (2)). This enables the detailed investigation of the modification dynamics in vivo . The addition of labeled methionine , either in medium (1) or medium (2), allows the observation of methylation processes. Other isotope-labeled metabolites may allow further modification analysis.
Overall, NAIL-MS enables the investigation of the RNA modification dynamics by means of mass spectrometry. Using this technique, enzymatic demethylation has been observed for multiple RNA damage within living bacteria.
Discovery of new RNA modifications
For the discovery of uncharacterized modifications, cells are grown in unlabeled, 13 C-labeled, 15 N-labeled, 2 H-labeled or 34 S-labeled medium. Unknown signals that occur during mass spectrometry are then examined in all differently labeled cultures. If retention times of unknown compounds with correspondingly diverging mass-to-charge ratios overlap, a molecular formula for the compound can be predicted by calculating the mass differences of the overlapping signals in the differently labeled cultures. With this method several new RNA modifications could be discovered. This experimental design was also the original idea with which the concept of NAIL-MS began.
Oligonucleotide NAIL-MS
NAIL-MS can also be used for oligonucleotide analysis using mass spectrometry. This is useful when you want to keep the sequence information.
Web links
Individual evidence
- ↑ Valentin F. Reichle, Steffen Kaiser, Matthias Heiss, Felix Hagelskamp, Kayla Borland: Surpassing limits of static RNA modification analysis with dynamic NAIL-MS . In: Methods . tape 156 , March 1, 2019, p. 91-101 , doi : 10.1016 / j.ymeth.2018.10.025 .
- ↑ Stefanie Kellner, Jennifer Neumann, David Rosenkranz, Svetlana Lebedeva, René F. Ketting: Profiling of RNA modifications by multiplexed stable isotope labeling . In: Chemical Communications . tape 50 , no. 26 , April 4, 2014, ISSN 1359-7345 , p. 3516 , doi : 10.1039 / c3cc49114e .
- ↑ a b Valentin F. Reichle, Dimitar P. Petrov, Verena Weber, Kirsten Jung, Stefanie Kellner: NAIL-MS reveals the repair of 2-methylthiocytidine by AlkB in E. coli . In: Nature Communications . tape 10 , no. 1 , December 2019, ISSN 2041-1723 , p. 5600 , doi : 10.1038 / s41467-019-13565-9 , PMID 31811240 , PMC 6898146 (free full text).
- ↑ Christina Dal Magro, Patrick Keller, Annika Kotter, Stephan Werner, Victor Duarte: A Vastly Increased Chemical Variety of RNA Modifications Containing a Thioacetal Structure . In: Angewandte Chemie International Edition . tape 57 , no. 26 , June 25, 2018, p. 7893–7897 , doi : 10.1002 / anie.201713188 .
- ↑ Eoin P. Quinlivan, Jesse F. Gregory: DNA digestion to deoxyribonucleoside: A simplified one-step procedure . In: Analytical Biochemistry . tape 373 , no. 2 , February 15, 2008, p. 383-385 , doi : 10.1016 / j.ab.2007.09.031 , PMID 18028864 , PMC 2239294 (free full text).
- ↑ Pamela F. Crain: [42] Preparation and enzymatic hydrolysis of DNA and RNA for mass spectrometry . In: Methods in Enzymology . tape 193 . Elsevier, 1990, ISBN 978-0-12-182094-7 , pp. 782-790 , doi : 10.1016 / 0076-6879 (90) 93450-y .
- ↑ a b Valentin F. Reichle, Verena Weber, Stefanie Kellner: NAIL ‐ MS in E. coli Determines the Source and Fate of Methylation in tRNA . In: ChemBioChem . tape 19 , no. 24 , December 18, 2018, ISSN 1439-4227 , p. 2575-2583 , doi : 10.1002 / cbic.201800525 , PMID 30328661 , PMC 6582434 (free full text).
- ^ Matthias Heiss, Valentin F. Reichle, Stefanie Kellner: Observing the fate of tRNA and its modifications by nucleic acid isotope labeling mass spectrometry: NAIL-MS . In: RNA Biology . tape 14 , no. 9 , September 2, 2017, ISSN 1547-6286 , p. 1260–1268 , doi : 10.1080 / 15476286.2017.1325063 , PMID 28488916 , PMC 5699550 (free full text).
- ↑ Stefanie Kellner, Antonia Ochel, Kathrin Thüring, Felix Spenkuch, Jennifer Neumann: Absolute and relative quantification of RNA modifications via biosynthetic isotopomers . In: Nucleic Acids Research . tape 42 , no. 18 , October 13, 2014, ISSN 1362-4962 , p. e142 – e142 , doi : 10.1093 / nar / gku733 , PMID 25129236 , PMC 4191383 (free full text).
- ↑ Felix Hagelskamp, Kayla Borland, Jillian Ramos, Alan G Hendrick, Dragony Fu: Broadly applicable oligonucleotide mass spectrometry for the analysis of RNA writers and erasers in vitro . In: Nucleic Acids Research . tape 48 , no. 7 , April 17, 2020, ISSN 0305-1048 , p. e41 – e41 , doi : 10.1093 / nar / gkaa091 , PMID 32083657 , PMC 7144906 (free full text).