Endogenous retrovirus
Endogenous retroviruses ( ERV ) are retroviruses that do not go through a full replication cycle , but are passed on as a provirus from generation to generation in the host's genome . It is believed that they originated many generations ago through infection of the germ line cells of humans and other vertebrates .
properties
Retroviruses are viruses that use the viral enzyme reverse transcriptase to transcribe their RNA genome in DNA so that they can be integrated into the genome of their host cell. Most retroviruses can only infect a few somatic cell types . If some succeed in infecting germ cells and thus being passed on to subsequent generations, they become endogenous retroviruses that can remain in the genome of their host species for long periods of time.
In some cases, endogenous retroviruses only remain infectious for a short time (a few hundred generations) because mutations accumulate during replication by the host, which lead to creeping virus inactivation. Basically, the endogenous retroviruses tend to mutations because they do not code for proteins that are necessary for the cells.
Others remain active and can continue to produce exogenous virus particles . Complete proviruses that are in principle replicable , however, are the exception and only make up around 0.5 percent of the human genome.
Under certain conditions, the endogenous retroviruses can be remobilized, which then carry out a transposition . These mobile elements, in which only the LTR regions are partially still present ( conserved ), are then also referred to as LTR retrotransposons .
history
Endogenous retroviruses were discovered in the late 1960s. Three different types of endogenous retroviruses have been described at about the same time: the avian leukosis virus (ALV) from domestic fowl ( Gallus gallus ) and the murine leukemia virus (MLV) and the mouse mammary tumor virus (MMTV) from domestic mice ( Mus musculus ).
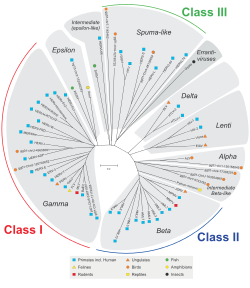
Until 2007 it was assumed that only the simple, but not the complex retroviruses can become endogenous retroviruses (the spumaviruses are an exception ). In 2007, the first endogenous retrovirus that originates from a lentivirus and thus from a complex retrovirus was described: the rabbit endogenous lentivirus type K (RELIK). The only retrovirus genus for which no endogenous retroviruses have been described so far are the delta retroviruses.
Human endogenous retroviruses
Human endogenous retroviruses ( HERV ) are found in large numbers in the human genome. Since they became part of the genome a very long time ago, numerous mutations of all kinds have accumulated in their sequence, including point mutations , deletions , insertions of other retro elements , recombinations, and mini and microsatellite expansion. The retroviral sequences are therefore already heavily modified and often difficult to find. Some HERVs are suspected to be involved in the development of some autoimmune diseases , particularly multiple sclerosis . Others take care of the development and regulation of important organs, e.g. B. the placenta in mammals.
As part of the Human Genome Project in the human genome thousands of ERV were found, the first in 24 different "families" are divided and account for about 8% of the human genome. According to more recent findings, there are already 31 different groups, each of which has arisen through a single integration event.
Endogenous retroviruses in pigs
Porcine endogenous retroviruses ( PERV ) are the endogenous retroviruses found in pigs . Depending on the breed of pig, there are 3-140 copies of PERV in the pig's genome. The retroviruses PERV-A and PERV-B are found in all pig breeds, PERV-C in most. The first two forms can also infect human cells in laboratory tests, PERV-C only pig cells. However, recombinations of the PERV can form, which can then also be infectious.
The only way to inactivate them is genome removal using genome editing . An earlier attempt with zinc finger nucleases failed because the multiple gene changes ultimately turned out to be cytotoxic . However, it has now been possible to eliminate 62 PERV sequences in immortal pig cell lines using CRISPR-Cas , and in 2017 living pigs were bred after 25 PERV sequences had been removed from the genome.
Research is focused on PERV and its inactivation, because pigs can be used as organ donors for xenotransplantations and PERV represents a safety risk.
literature
- Retrotransposons, Endogenous Retroviruses, and the Evolution of Retroelements. In: John M. Coffin, Stephen H. Hughes, Harold Varmus: Retroviruses. Cold Spring Harbor Laboratory Press, Plainview NY 1997, ISBN 0-87969-497-1 .
- Roswitha Löwer, Johannes Löwer , Reinhard Kurth : The viruses in all of us: Characteristics and biological significance of human endogenous retrovirus sequences. In: Proceedings of the National Academy of Sciences. (PNAS) May 28, 1996, Vol. 93, No. 11, pp. 5177-5184, full text (PDF).
- Luis P Villarreal: Viruses and the Evolution of Life. ASM Press, Washington 2005.
Web links
- kereng: Evidence of evolution through endogenous retroviruses , AG EvoBio from September 13, 2010, accessed on February 16, 2020.
- HERVd - Human Endogenous Retrovirus Database , Institute of Molecular Genetics of the ASCR, accessed February 16, 2020.
- Nadja Podbregar: Secret Helpers - What Function Do Endogenous Retroviruses Have in Us? , on: scinexx.de from November 5, 2010, accessed on February 16, 2020
- Reinhard Kurth : Discovery of viruses in the human genome , Spectrum of Science 9/1993
Individual evidence
- ↑ Christiane Hohmann: Parasites in the genome. In: Pharmaceutical newspaper . July 2010, accessed August 11, 2017 .
- ^ Robin A. Weiss: The discovery of endogenous retroviruses In: Retrovirology. October 3, 2006, Volume 3, p. 67 → Review. PMID 17018135 , doi: 10.1186 / 1742-4690-3-67 .
- ↑ Aris Katzourakis, Michael Tristem, Oliver G. Pybus, Robert J. Gifford: Discovery and analysis of the first endogenous lentivirus. In: Proceedings of the National Academy of Sciences. (PNAS) April 10, 2007, Volume 104, No. 15, pp. 6261-6265, PMID 17384150 , full text
- ↑ Jan Paces, Adam Pavlícek, Václav Paces: HERVd: database of human endogenous retroviruses. In: Nucleic Acids Research . 2002, Vol. 30, No. 1, pp. 205-206, full text.
- ↑ P. Perot, PA Bolze, F. Mallet: From Viruses to Genes: Syncytins. In: Günther Witzany (Ed.): Viruses: Essential Agents of Life. Springer, Dordrecht / New York 2012, ISBN 978-94-007-4898-9 , pp. 325–361.
- ↑ E. Lander et al. a .: Initial sequencing and analysis of the human genome. In: Nature . 2001, Vol. 409, pp. 860-921.
- ↑ EC Holmes: Ancient lentiviruses leave their mark. In: Proceedings of the National Academy of Sciences . (PNAS) April 10, 2007, Vol. 104, No. 15, pp. 6095-6096, PMID 17404211 .
- ^ RJ Gifford: Evolution at the host-retrovirus interface. In: Bioessays. Dec. 2006, Vol. 28, No. 12, pp. 1153-1156, PMID 17117481 .
- ↑ Joachim Denner: Paving the path toward procine Organs for transplantation New England Journal of Medicine 2017, Volume 377, Issue 19 of November 9, 2017, pages 1891-1893, doi: 10.1056 / NEJMcibr1710853
- ↑ Sven Stockrahm: Pig is my whole heart. In: Zeit Online . July 11, 2017. Retrieved August 14, 2017 .