Rotavirus
| Rotavirus | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|

Rotaviruses in a TEM image (length scale 100 nm) |
||||||||||||||||||
| Systematics | ||||||||||||||||||
|
||||||||||||||||||
| Taxonomic characteristics | ||||||||||||||||||
|
||||||||||||||||||
| Scientific name | ||||||||||||||||||
| Rotavirus | ||||||||||||||||||
| Left | ||||||||||||||||||
|
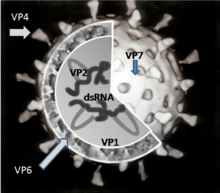
The genus Rotavirus includes viruses of the subfamily Sedoreovirinae in the family Reoviridae with a genome of eleven segments of a double-stranded RNA (dsRNA). Rotaviruses have very complex virus particles (virions), which consist of two concentric capsids and an inner core structure; this is also referred to as a three-layer capsid structure. Protein bridges and channels can be seen in TEM images between the outer and inner capsid , which appear to be wheel-like in the negative contrast. The name of the genus (from Latin rota "wheel") is derived from this appearance . Rotaviruses were first identified as causing diarrhea in mice in the 1950s, and have since been found in several mammals and birds. For humans, subtypes from three rotavirus species are known, which are grouped together as human rotaviruses .
Systematics
The viruses of the genera Coltivirus and Orbivirus (the latter, for example, with the species bluetongue virus ), both in the same subfamily Sedoreovirinae , also have a similar structure with two concentric shells .
The genus Rotavirus is currently divided into 9 species (AI). One of the most important criteria for defining a rotavirus species is that reassortment can only take place between the subtypes , but not between individual species. They thus form a uniform gene pool that can create new subtypes by means of antigen shift and antigen drift . When the sequence of the coreprotein VP2 is compared, the species rotavirus A and C appear to be closely related, while rotavirus B has significant sequence differences.
- Genus Rotavirus
-
- Species Rotavirus A
- Subtype Simian rotavirus A
- Avian rotavirus A subtype
- Bovine rotavirus A subtype
- Subtype porcine rotavirus A
- Subtype caprine rotavirus A
- Equines rotavirus A subtype
- Subtype Human Rotavirus A
- Subtype Lapines Rotavirus A
- Rabbit rotavirus subtype
- Preliminary subtypes of the species:
- Rotavirus subtype of lambs
- Canine rotavirus subtype
- Rhesus rotavirus subtype
-
- Species Rotavirus B
- Subtype (serogroup) bovine rotavirus B
- Subtype human rotavirus B
-
- Species Rotavirus C
- Subtype human rotavirus C
-
- Species Rotavirus D
- Chicken rotavirus D subtype
-
- Species Rotavirus E
- Subtype porcine rotavirus E
-
- Species Rotavirus F
- Chicken rotavirus F subtype
-
- Species Rotavirus G
- Chicken rotavirus G subtype
- Species Rotavirus H
- Species Rotavirus I
swell
- RF Ramig et al .: Genus Rotavirus . In: CM Fauquet, MA Mayo et al .: Eighth Report of the International Committee on Taxonomy of Viruses . London, San Diego 2005, ISBN 0-12-249951-4 , pp. 484-496
- Robert D. Shaw and Harry B. Greenberg: Rotaviruses (Reoviridae) . In: Allan Granoff, Robert G. Webster (eds.): Encyclopedia of Virology . San Diego, 1999 ( Vols 1-3), ISBN 0-12-227030-4 , Vol 3, pp 1576-1592
- J. Matthijnssens et al .: Full genomic analysis of human rotavirus strain B4106 and lapine rotavirus strain 30/96 provides evidence for interspecies transmission . J. Virol. (2006) 80 (8): pp. 3801-3810, PMID 16571797
- JA Lawton et al .: Three-dimensional structural analysis of recombinant rotavirus-like particles with intact and amino-terminal-deleted VP2: implications for the architecture of the VP2 capsid layer . J. Virol. (1997) 71 (10): pp. 7353-7360, PMID 9311813 , PMC 192080 (free full text)
Web links
- Genus Rotavirus (NCBI)
- Rotavirus in the ICTV database
- Early Steps of Viral Assembly Revealed by Genetically Engineered Slow-Growing Mutant Rotavirus , on: SciTechDaily from June 25, 2020, Source: Baylor College of Medicine
- S. Lopez, C. F. Arias: Early Steps in Rotavirus Cell Entry , in: Curr Top Microbiol Immunol 309, 2006, pp. 39-66, doi: 10.1007 / 3-540-30773-7_2 , PMID 16909896 .
Individual evidence
- ↑ ICTV Master Species List 2018b v1 MSL # 34, Feb. 2019
- ↑ a b c d e ICTV: Bluetongue virus , EC 51, Berlin, Germany, July 2019; Email ratification March 2020 (MSL # 35)
- ↑ Cornelia Henke-Gendo: Viral gastroenteritis pathogens . In: Sebastian Suerbaum, Gerd-Dieter Burchard, Stefan HEKaufmann, Thomas F. Schulz (eds.): Medical microbiology and infectious diseases . Springer-Verlag, 2016, ISBN 978-3-662-48678-8 , pp. 513 ff ., doi : 10.1007 / 978-3-662-48678-8_65 .
- ↑ Attoui H, Mohd Jaafar F, de Micco P, de Lamballerie X: Coltiviruses and seadornaviruses in North America, Europe, and Asia . In: Emerg Infect Dis . 11 (11), November 2005, pp. 1673-1679. doi : 10.3201 / eid1111.050868 . PMID 16318717 . PMC 3367365 (free full text).
- ↑ ICTV : Master Species List 2018a v1 , MSL including all taxa updates since the 2017 release. Fall 2018 (MSL # 33)