Single guide RNA
Single Guide RNA (sgRNA) is an artificial RNA that is used in the CRISPR / Cas method , CRISPRi or CRISPRa in combination with Cas9 or Cas12b .
properties
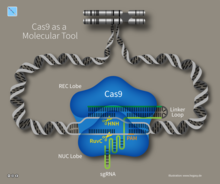
The sgRNA forms a secondary structure called an R-loop . It can be used in bacteria , yeast , fruit flies , zebrafish, and mice . It is then bound by Cas proteins of type I and II. Cas9 naturally binds two RNAs, the crRNA and the tracrRNA , while in the method only one sgRNA consisting of sequences of the two RNAs is used. This means that only one RNA has to be cloned for the CRISPR / Cas method . An sgRNA consists of the 20 nucleotides upstream (in the 5 'direction) of a Protospacer Adjacent Motif (PAM) of the target DNA to be cut and part of the tracrRNA. There is preferably a guanine nucleotide ( GC clamp ) at the 5 'end of the 20 nucleotides (position 1) and an adenine or thymine nucleotide four nucleotides in front of the PAM (position 17) . Programs for identifying 20 nucleotides in front of a PAM or for designing an sgRNA are, for example, CHOPCHOP, CasOFFinder, FlyCRISPR, CRISPR-ERA, SgRNA Designer, CRISPOR, E-CRISP and CRISPRdirect.
In most cases, viral vectors are transduced or plasmids are transfected to introduce the sgRNA into eukaryotic cells . When used for gene therapy in eukaryotes, a eukaryotic promoter is used. If the Cas protein and the sgRNA are introduced into a cell , the sgRNA is generated beforehand by in vitro transcription from a vector, for example with a T7 promoter .
Web links
- How to Design Your gRNA for CRISPR Genome Editing , accessed January 29, 2019.
Individual evidence
- ^ A b F. Jiang, DW Taylor, JS Chen, JE Kornfeld, K. Zhou, AJ Thompson, E. Nogales, JA Doudna: Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. In: Science . Volume 351, number 6275, February 2016, pp. 867-871, doi : 10.1126 / science.aad8282 , PMID 26841432 , PMC 5111852 (free full text).
- ↑ W. Jiang, D. Bikard, D. Cox, F. Zhang, LA Marraffini: RNA-guided editing of bacterial genomes using CRISPR-Cas systems. In: Nature Biotechnology . Volume 31, number 3, March 2013, pp. 233-239, doi : 10.1038 / nbt.2508 , PMID 23360965 , PMC 3748948 (free full text).
- ↑ JM Peters, A. Colavin, H. Shi, TL Czarny, MH Larson, S. Wong, JS Hawkins, CH Lu, BM Koo, E. Marta, AL Shiver, EH Whitehead, JS Weissman, ED Brown, LS Qi, KC Huang, CA Gross: A Comprehensive, CRISPR-based Functional Analysis of Essential Genes in Bacteria. In: Cell . Volume 165, number 6, June 2016, pp. 1493–1506, doi : 10.1016 / j.cell.2016.05.003 , PMID 27238023 , PMC 4894308 (free full text).
- ↑ XT Li, Y. Jun, MJ Erickstad, SD Brown, A. Parks, DL Court, S. Jun: tCRISPRi: tunable and reversible, one-step control of gene expression. In: Scientific Reports . Volume 6, 12 2016, p. 39076, doi : 10.1038 / srep39076 , PMID 27996021 , PMC 5171832 (free full text).
- ^ JE DiCarlo, JE Norville, P. Mali, X. Rios, J. Aach, GM Church: Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. In: Nucleic acids research. Volume 41, number 7, April 2013, pp. 4336-4343, doi : 10.1093 / nar / gkt135 , PMID 23460208 , PMC 3627607 (free full text).
- ↑ GW Thickbroom, FL Mastaglia: Cerebral events preceding- self-paced and visually triggered saccades. A study of presaccadic potentials. In: Electroencephalography and clinical neurophysiology. Volume 62, Number 4, July 1985, pp. 277-289, doi : 10.1016 / 0168-5597 (85) 90005-x , PMID 2408874 .
- ↑ WY Hwang, Y. Fu, D. Reyon, ML Maeder, SQ Tsai, JD Sander, RT Peterson, JR Yeh, JK Joung: Efficient genome editing in zebrafish using a CRISPR-Cas system. In: Nature Biotechnology . Volume 31, number 3, March 2013, pp. 227-229, doi : 10.1038 / nbt.2501 , PMID 23360964 , PMC 3686313 (free full text).
- ↑ H. Wang, H. Yang, CS Shivalila, MM Dawlaty, AW Cheng, F. Zhang, R. Jaenisch: One-step generation of mice carrying mutations in multiple genes by CRISPR / Cas-mediated genome engineering. In: Cell . Volume 153, number 4, May 2013, pp. 910-918, doi : 10.1016 / j.cell.2013.04.025 , PMID 23643243 , PMC 3969854 (free full text).
- ↑ M. Jinek, K. Chylinski, I. Fonfara, M. Hauer, JA Doudna, E. Charpentier: A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. In: Science . Volume 337, number 6096, August 2012, pp. 816-821, doi : 10.1126 / science.1225829 , PMID 22745249 , PMC 6286148 (free full text).
- ↑ How to design sgRNA sequences. In: takarabio.com. Retrieved February 2, 2019 .
- ↑ CHOPCHOP. In: chopchop.cbu.uib.no. Retrieved January 30, 2019 .
- ↑ CRISPR RGEN Tools. In: rgenome.net. Retrieved January 30, 2019 .
- ^ C. Dustin Rubinstein, Ed O'Connor-Giles, Kate O': CRISPR Optimal Target Finder. In: tools.flycrispr.molbio.wisc.edu. Retrieved January 30, 2019 .
- ↑ CRISPR-ERA. In: crispr-era.stanford.edu. Accessed January 30, 2019 .
- ↑ sgRNA Designer: CRISPRko. In: portals.broadinstitute.org. Retrieved January 30, 2019 .
- ↑ CRISPOR. In: crispor.tefor.net. Accessed January 30, 2019 .
- ↑ E-CRISP design. In: e-crisp.org. January 1, 2013, accessed January 30, 2019 (enc).
- ^ SE Mohr, Y. Hu, B. Ewen-Campen, BE Housden, R. Viswanatha, N. Perrimon: CRISPR guide RNA design for research applications. In: The FEBS journal. Volume 283, number 17, 09 2016, pp. 3232–3238, doi : 10.1111 / febs.13777 , PMID 27276584 , PMC 5014588 (free full text).