Segmentation gene
Segmentation genes determine during embryogenesis of insects , the number and internal organization of the segments . They were researched on the model organism Drosophila melanogaster , primarily by analyzing the genes of mutated flies or their embryos , which showed malformations of the segmentation or body structure. The genes have names that are usually derived from mutations that led to their discovery. The gene product of the segmentation genes are proteins with regulatory tasks that attach to the DNA and thus switch other genes on and off like a switch, these are called transcription factors . For this purpose, the target genes have sequences which are arranged on the DNA strand in front of the protein-coding gene segment and which are not themselves transcribed . Since the front end of a DNA strand is referred to as the "cis" end (the rear end as "trans"), one speaks of "cis-regulatory" sections or cis elements . The transcription factors of the segmentation genes are connected in series in a regulatory cascade, which means that higher-level, early segmentation genes switch the later ones on or off depending on their location in the developing embryo. A striped pattern develops from strips arranged one behind the other, in each of which a certain segmentation gene (or a group of such) is active. The later segmentation of the body is represented by these cell strips. The cells that build the body tissue themselves receive information about their location in the developing organism from the segmentation genes. Depending on their location, they can grow, divide and differentiate or even die ( programmed cell death or apoptosis).
Later analyzes have shown that homologous genes of numerous of the segmentation genes occur in the entire animal kingdom in all of the organisms examined, they organize the formation of the body axes everywhere. This is done in a very similar and analogous way in non-segmented organisms. Other segmentation genes are only insects or arthropods pronounced, some even only in the Diptera (Diptera), individual are even exclusively from Drosophila known.
The regulation cascade
The embryo of the fruit fly Drosophila develops from the egg cell in that an outer layer of cells, the blastoderm , grows around a central reservoir of yolk . A germ line differentiates within the blastoderm . In the first steps of division, only the cell nuclei divide, while the cytoplasm is not divided by cell membranes ; such an undivided association is called a syncytium . Morphologically , no structure can be recognized at this stage; the front and rear ends of the embryo that are created look the same. Experiments in the 1960s and 1970s showed that the longitudinal axis of the body is already fixed at this stage. It turned out that the organizing factor that determines the longitudinal axis of the body is already given to the egg by the mother. This is based on substances that are concentrated in the anterior and posterior egg poles. Mutations in these genes lead to severe malformations, e.g. B. the rear half is created twice instead of the front half of the body. Since the corresponding gene products (transcripts) come from the mother, the corresponding genes are called maternal genes . The products of the maternal genes transferred into the cytoplasm of the egg determine the front and rear ends of the embryo. Their proteins form a concentration gradient in the embryo , with some at the front end in the highest concentration and some at the rear end. The cells in between contain different proportions of the two, depending on how far in front or behind they are. Depending on the concentration of the proteins in the maternal genes, a group of other genes is activated in the embryo , which are called gap genes (more often, taken from English, gap genes). The naming is based on the fact that when one of these genes is mutated, entire body sections are missing in the embryo. In Drosophila are five Gap genes. Each gap gene now activates, depending on its location, a different gene of a further class, the pair rule genes . The name is based on the fact that in the event of a mutation the embryo only has half the number of segments. This organizes the embryo into seven strips. The next class of pair rule genes activates the segment polarization genes . Each strip is divided into two partial strips. The fourteen body segments of the fly maggot are thus pre-formed. (In fact, the matter is a little more complicated: So-called parasegments are created, each consisting of the front end of one segment and the rear end of the adjacent segment).
In the finished organism, however, these segments are not morphologically the same, but differentiate into body sections ( Tagmata ): head, trunk ( thorax ) and abdomen ( abdomen ). Depending on the location, different attachments and other organs such as antennas , legs, etc. are formed or not. This identity is assigned to the segments by another class of genes , the Hox genes . There are eight Hox genes in the fruit fly. The expression of the Hox genes is not linked precisely to the sequence of segments. Some body segments express the same Hox gene, while others express two of them.
A total of about 40 to 50 genes have been identified that are involved in this pattern formation, the role of some of them is still unclear. A similar signal sequence, in which maternal and embryonic genes are also involved, defines the dorsal-ventral axis of the embryo, i.e. H. determined above (back side) and below (stomach side). These genes were discovered later and generally less well known.
Maternal genes
As the name suggests, the maternal genes are active in the maternal organism. Their gene products, mostly RNA ( transcripts ), more rarely finished proteins, are given to the developing egg. The genes Caudal (cad) and Hunchback (hb) have a dual role; they are transcribed both maternally and later in the embryo itself. The most important of the maternal genes that determine the orientation of the longitudinal axis of the body is bicoid (bcd). The bicoid protein defines the front end of the embryo and, depending on the concentration, activates other genes in zones of different widths progressing backwards. Cad and Hb are initially almost equally distributed in the egg. But since Bcd inhibits their expression, they accumulate in the rear area. Another group of RNAs and proteins is concentrated in the posterior egg pole.
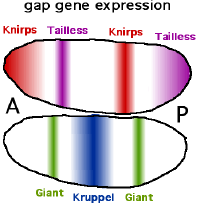
Gap genes
The name Lückengen ( Engl. Therefore, Gap gene) is because a malfunction of these genes cause gaps in segmentation, lack of body segments leads. You are responsible for the division into a front, middle and rear area. The currently eleven known gap genes include Giant (gt or gat), Hunchback (hb), Knirps (kni), Krüppel (mostly "kruppel") (kr) and Tailless (tll). cad and hb have a dual role. They are expressed both maternally (of maternal origin) and in the embryo itself. The gap genes are initially expressed in a relatively widely distributed manner and are later restricted to the appropriate strips through self-organization . Like the products of the maternal genes, the expressed transcription factors are only active for a short time. Your activity ends as soon as the pattern has developed and the following steps have been initiated, which is the case after around two hours of development. They no longer play a role in the later organism, but in some cases they are involved in further pattern formation processes regardless of their role in determining the body axis.
The expression of the gap genes is regulated by a combination of maternal genes and interactions between the gap genes. The torso (tor) gene, which is expressed at the cell ends but not in between, also plays a role. Torso encodes a transmembrane receptor that is activated by substances in the egg shell. The maternal Bcd protein switches on the hb gene in the front half of the body, so that two sharply divided halves are created (all-or-nothing reaction). At the same time, Bcd suppresses the transcription of cad, so that the Cad protein is only found in the back of the body. The other gap genes similarly form one or two stripes in different parts of the embryo. Kr is mainly formed in a region near the midline, where it is activated by Bcd but repressed (prevented) by Hb . Kni is expressed by a similar mechanism at the anterior end and in a stripe in the posterior portion.
The different parts of the body are usually characterized by one or a combination of two gap proteins. As a rule, the gap proteins show concentrations that rise to a maximum and then fall behind, the areas of which overlap to a greater or lesser extent.
Pair rule genes
The embryonic stripes that are given by the gap genes are each specified by different combinations of the pair rule genes. The pair of control genes Even (eve) and Fushi-tarazu (ftz) are respectively alternately in seven strips expressed . Other genes like Runt and Hairy show similar patterns. The aperiodic stripe pattern of the gap genes is overlaid by a periodic pattern. In the final state, the concentrations of the pair rule proteins are sharply differentiated depending on the cell position; they no longer overlap like those of the gap genes. Mutations in pair rule genes cause the loss of every second segment, so pair rule genes control the formation of the even-numbered or odd-numbered segments.
Segment polarity genes
The segment polarity genes determine both the final sequence of the (para-) segments and their polarity, i. H. her front and rear ends, solid. The segment polarity gene Engrailed (en) is expressed in a narrow zone near the front end of fourteen parasegment strips. Hedgehog (hh) shows a similar pattern. The Wingless (wn) gene, on the other hand, is active in strips near the rear end of the parasegmental strips. This pattern divides the germinal strips of the embryo into strips of the same type, which, however, are not generated synchronously but one after the other. In contrast to the preceding segmentation genes, the segment polarity genes remain active for a long time, in the case of engrailed up to the winged insect ( imago ). With the activity of the segment polarity genes, the segmentation is complete. In the following stages of development, the identity of the various segments is further specified. This is primarily the job of the Hox genes. In the activation of the specific Hox genes, the gap genes (which, in contrast to the later stages of the segmentation cascade, are not periodic) directly play an important role.
Occurs in other segmented animals
The basic scheme that was discovered in the model organism Drosophila was found in later research in the main features of all the arthropods examined. In all species, the segment formation takes place via a regulation cascade, via a germ band divided into segments by the segment polarity genes. The previous stages of development differ in detail in other arthropods. This is e.g. For example, unlike Drosophila, most arthropods do not begin their development with a syncytium. In addition, in a large number of species, the segments are not created at the same time, but only gradually formed in the course of ontogenesis at a segment formation zone at the rear end. In the relatively few species that have been investigated in more detail so far, bicoid (or a homologous gene) could only be found in other two-winged animals (dipteras). In other species, maternal genes are also involved in the structure, but different in detail. The genes cad and nanos (nos) seem to be widespread. Orthologous or homologous genes to most Drosophila segmentation genes have been found in most arthropods, but their role appears to be different in detail. In addition, other regulatory patterns were found in arachnids and millipedes , which are more similar to the formation of somites in vertebrates .
literature
- Michael Akam (1987): The molecular basis for metameric pattern in the Drosophila embryo. Development 101: 1-22.
- Dmitri Papatsenko, Michael Levine (2011): The Drosophila Gap Gene Network Is Composed of Two Parallel Toggle Switches. PLoS ONE 6 (7): e21145. doi : 10.1371 / journal.pone.0021145
- Andrew D. Peel, Ariel D. Chipman, Michael Akam (2005): Arthropod segmentation: beyond the Drosophila paradigm. Nature Review Genetics doi : 10.1038 / nrg1724
Individual evidence
- ↑ CR Bartram et al. : Human genetic diagnostics: Scientific principles and social consequences. Springer, 2000, ISBN 978-3540679455 , p. 30.
- ↑ a b January Zravý, David Storch, Stanislav Mihulka: Evolution: A reading textbook. Spektrum Akademischer Verlag, 2009, ISBN 978-3827419750 , p. 230.
- ↑ Monica Hirsch-Kauffmann, Manfred Schweiger: Biology and molecular medicine for physicians and natural scientists. Thieme Verlag, 2009, ISBN 978-3137065074 , p. 254.