Phytoene desaturase
| Vegetable phytoene desaturase | ||
|---|---|---|
|
||
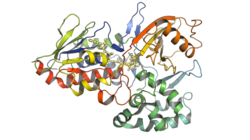
| Crystallographic structure of phytoene desaturase from rice. | ||
| Mass / length primary structure | 491 amino acids | |
| Cofactor | FAD | |
| Identifier | ||
| Gene name (s) | PDS | |
| External IDs | ||
| Enzyme classification | ||
| EC, category | 1.3.5.5 , oxidoreductase | |
| Response type | Dehydrogenation | |
| Substrate | 15 cis phytoene | |
| Products | 9,15,9'-tri-cis-ζ-carotene | |
| Occurrence | ||
| Parent taxon | plants | |
| Bacterial phytoene desaturase | ||
|---|---|---|

|
||
| Crystallographic structure of the phytoene desaturase from Pantoea ananatis. | ||
| Mass / length primary structure | 501 amino acids | |
| Identifier | ||
| Gene name (s) | CRTI | |
| External IDs | ||
| Enzyme classification | ||
| EC, category | 1.3.99.31 , oxidoreductase | |
| Response type | Dehydrogenation | |
| Substrate | 15 cis phytoene | |
| Products | all- trans -z-lycopene | |
| Occurrence | ||
| Parent taxon | bacteria | |
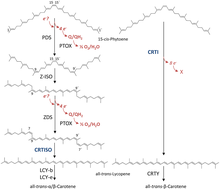
Phytoene desaturases are at the carotenoid - biosynthesis involved enzymes from the group of desaturases . They catalyze the dehydrogenation of 15-cis- phytoene . A distinction is made between bacterial and fungal phytoene desaturases (CRTI) and plant and cyanobacterial phytoene desaturases (PDS). CRTI catalyzes the conversion of phytoene directly to all-trans- lycopene . The complete conversion in plants and bacteria takes place via four enzymes (PDS, ζ-carotene isomerase, ζ-carotene desaturase, cis-trans isomerase), of which PDS catalyzes the first step to ζ-carotene .
biochemistry
When 15-cis-phytoene is converted into all-trans-lycopene, a total of four new double bonds are introduced into the substrate molecule. The oxidation of the phytoene takes place with the reduction of FAD , which is contained in both bacterial and plant phytoene desaturases. While CRTI represents the entire biochemical pathway to lycopene, only the catalysis 9,15,9' tri done at PDS cis -ζ-carotene. Two new double bonds are introduced at position 11 and 11 'of the symmetrical carbon chain and the double bonds are isomerized from trans to cis at positions 9 and 9' . The electrons removed in this reaction are carried away to plastoquinone via FAD and ultimately used for oxygen reduction via terminal oxidases.
use
Some herbicides such as norflurazon , diflufenican and picolinafen work by inhibiting PDS. The herbicide blocks the plastoquinone binding site within the enzyme. Since switching off the PDS gene in plants causes the plant to bleach , this effect was used to present efficient genome editing using CRISPR / Cas9 .
In the golden rice , in addition to a phytoene synthase and lycopene cyclase from the narcissus, the gene of a bacterial phytoene desaturase was integrated into the rice genome.
Individual evidence
- ↑ a b c Anton Brausemann, Sandra Gemmecker, Julian Koschmieder, Sandro Ghisla, Peter Beyer, Oliver Einsle: Structure of Phytoene Desaturase Provides Insights into Herbicide Binding and Reaction Mechanisms Involved in Carotene Desaturation . In: Structure . tape 25 , no. 8 , 2017, p. 1222–1232.E3 , doi : 10.1016 / j.str.2017.06.002 , PMID 28669634 .
- ↑ Patrick Schaub, Qiuju Yu, Sandra Gemmecker, Pierre Poussin-Courmontagne, Justine Mailliot, Alastair G. McEwen, Sandro Ghisla, Salim Al-Babili, Jean Cavarelli, Peter Beyer: On the Structure and Function of the Phytoene Desaturase CRTI from Pantoea ananatis , a Membrane-Peripheral and FAD-Dependent Oxidase / Isomerase . In: PLoS One . tape 7 , no. 6 , 2012, p. e39550 , doi : 10.1371 / journal.pone.0039550 , PMID 22745782 , PMC 3382138 (free full text).
- ↑ PD Fraser, N. Misawa, H. Linden, S. Yamano, K. Kobayashi: Expression in Escherichia coli, purification, and reactivation of the recombinant Erwinia uredovora phytoene desaturase . In: The Journal of Biological Chemistry . tape 267 , no. 28 , October 5, 1992, ISSN 0021-9258 , pp. 19891-19895 , PMID 1400305 .
- ↑ a b Alexander R. Moise, Salim Al-Babili, Eleanore T. Wurtzel: Mechanistic aspects of carotenoid biosynthesis . In: Chemical Reviews . tape 114 , no. 1 , January 8, 2014, ISSN 1520-6890 , p. 164-193 , doi : 10.1021 / cr400106y , PMID 24175570 .
- ↑ PD Fraser, H Linden, G Sandmann: Purification and reactivation of recombinant Synechococcus phytoene desaturase from an overexpressing strain of Escherichia coli. In: Biochemical Journal . tape 291 , no. 3 , May 1, 1993, ISSN 0264-6021 , p. 687-692 , PMID 8489496 .
- ↑ TA Dailey, HA Dailey: Identification of an FAD superfamily containing protoporphyrinogen oxidases, monoamine oxidases, and phytoene desaturase. Expression and characterization of phytoene desaturase of Myxococcus xanthus . In: The Journal of Biological Chemistry . tape 273 , no. 22 , May 29, 1998, ISSN 0021-9258 , pp. 13658-13662 , PMID 9593705 .
- ↑ SR Norris, TR Barrette, D. DellaPenna: Genetic dissection of carotenoid synthesis in arabidopsis defines plastoquinone as an essential component of phytoene desaturation . In: The Plant Cell . tape 7 , no. December 12 , 1995, ISSN 1040-4651 , pp. 2139-2149 , doi : 10.1105 / tpc.7.12.2139 , PMID 8718624 , PMC 161068 (free full text).
- ↑ Thomas Seitz, Michael G. Hoffmann, Hansjörg Krähmer: Herbicides for agriculture: Chemical weed control . In: ChiuZ . tape 37 , no. 2 , 2003, p. 118 , doi : 10.1002 / ciuz.200300279 .
- ↑ Genji Qin, Hongya Gu, Ligeng Ma, Yiben Peng, Xing Wang Deng: Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in Arabidopsis by impairing chlorophyll, carotenoid, and gibberellin biosynthesis . In: Cell Research . tape 17 , no. 5 , 2007, ISSN 1748-7838 , p. 471-482 , doi : 10.1038 / cr.2007.40 , PMID 17486124 .
- ↑ Chikako Nishitani, Narumi Hirai, Sadao Komori, Masato Wada, Kazuma Okada: Efficient Genome Editing in Apple Using a CRISPR / Cas9 system . In: Scientific Reports . tape 6 , 2016, ISSN 2045-2322 , p. 31481 , doi : 10.1038 / srep31481 , PMID 27530958 , PMC 4987624 (free full text).
- ↑ Ikuko Nakajima, Yusuke Ban, Akifumi Azuma, Noriyuki Onoue, Takaya Moriguchi: CRISPR / Cas9-mediated targeted mutagenesis in grape . In: PloS One . tape 12 , no. 5 , 2017, ISSN 1932-6203 , p. e0177966 , doi : 10.1371 / journal.pone.0177966 , PMID 28542349 , PMC 5436839 (free full text).
- ↑ Peter Schopfer: Plant Physiology . Elsevier, Spektrum Akademischer Verlag, Munich 2010, ISBN 978-3-8274-2351-1 , pp. 382-383 .