1q21.1 deletion syndrome
| Classification according to ICD-10 | |
|---|---|
| Q93.5 | Other deletions of a part of a chromosome |
| ICD-10 online (WHO version 2019) | |
The 1q21.1 deletion syndrome is a rare disease , which is obtained by a deletion in the human chromosome 1 at the point is caused 1q21.1. This change can result in mental retardation and various physical abnormalities. The penetrance and expressivity are variable. Some people with this deletion have no apparent impairment.
Unique, an international association of people with rare chromosomal abnormalities, knows 64 registered people with this deletion worldwide. (As of October 2012)
In addition to the 1q21.1 deletion, there is also a 1q21.1 duplication in which the section in question is present twice or three times.
Structure of the region 1q21.1
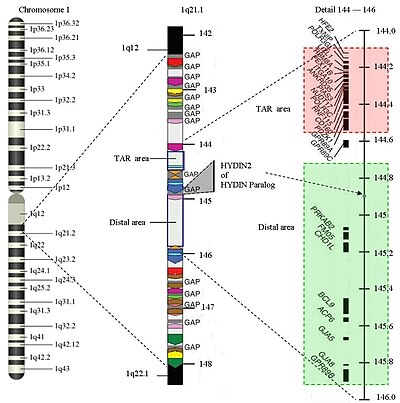
The structure of the 1q21.1 is complex. The area has a size of about 6 million base pairs (Mb) (from 141.5 to 147.9 Mb). There are two areas in which the deletion can occur: the TAR area, which results in TAR syndrome, and the distal area, which leads to other abnormalities. The area has several repetitions of the same structure (areas in the same color on the image have the same structure). Only 25% of the structure is specific. However, to date there is no complete information on the nucleotide sequence in these areas. Area 1q21.1 is considered to be one of the most difficult to map the human genome . The missing areas are currently around 700,000 base pairs. It is therefore difficult to pinpoint the start and end of a deletion.
Types
There are two types of 1q21.1 deletion syndrome:
- The so-called class I deletion is restricted to the TAR area or the distal area.
- If the deletion is so large that both areas are affected, it is referred to as a class II deletion. There are complex cases in which both the TAR and distal areas are affected while the area in between is normal, as well as some atypical variants.
A normal deletion affects between 1.0 and 1.9 million base pairs (Mb). According to Mefford, 1.35 Mb is the standard for such a deletion. The largest deletion observed in living humans is over 5 Mb.
Symptoms
The consequences of microdeletion are phenotypically very variable. While some people with this syndrome have no noticeable impairment, others have significant limitations. So far proven symptoms are:
- Haploinsufficiency
- TAR syndrome
- neurological - psychiatric problems: developmental retardation , mental retardation , autism , schizophrenia , epilepsy , atactic gait with a tendency to fall
- Dysmorphism : microcephaly , protruding forehead , bulbous nose, broad thumbs and toes, additional transverse fold of the fifth finger (class II deletion), hypermobility of the joints, pseudarthrosis of the collarbone (class II deletion), vagina laplasia ("Müllerian aplasia ")
- Cardiac abnormalities and cardiovascular abnormalities (30% of cases) such as an abnormal origin of the coronary arteries (Class II deletion)
- ophthalmic problems: cataracts , deep-set eyes , squint
- Kidney diseases : missing kidney, migrating kidney , neuroblastoma
The following symptoms could not yet be assigned to the 1q21.1 deletion syndrome with certainty:
- Backflow of stomach acid into the esophagus
- Non-compaction cardiomyopathy associated with Class II 1q21.1 deletion syndrome.
- increased neck transparency and oligohydramnios during pregnancy.
Since there is little information about the syndrome, the completeness of the above list of symptoms is uncertain.
Affected genes
The genes affected in 1q21.1 deletion syndrome in the TAR region are: HFE2 ( hemojuvelin ), TXNIP , POLR3GL , LIX1L , RBM8A , PEX11B , ITGA10 , ANKRD35 , PIAS3 , NUDT17 , POLR3C , RNF115 , CD160 , PDR89A and GPR89A1 .
The genes affected in the distal area are PDE4DIP , HYDIN2 , PRKAB2 , PDIA3P , FMO5 , CHD1L , BCL9 , ACP6 , GJA5 , GJA8 , NBPF10 , GPR89B , GPR89C , PDZK1P1 and NBPF11 .
Diagnosis
The syndrome can also occur in families in which neither parent carries the genes . Because of the repetitions in 1q21.1, there is a higher probability of an unequal crossing-over during meiosis . In this case, parts of the chromosome can be lost and copy number variation (CNV) occurs in the form of deletions or duplications. Such a random mutation is called a “ de novo ” situation and occurs in 75% of cases.
25% of the time, either parent is a carrier of the syndrome without affecting the person or with only mild symptoms. In several cases, the syndrome was first diagnosed in children because of autism or some other problem and it was only later that it was found that one of the parents was also affected.
In families where both parents tested negative for the syndrome, the risk of having a second child with the syndrome is extremely low. If the syndrome was found in the family, there is a 50% risk of the disease because the inheritance is autosomal dominant . However, the effects of the 1q21.1 microdeletion on the child cannot be predicted. Parents whose child has the syndrome should therefore be given genetic advice and examined prior to a further pregnancy .
A determination of the chromosome change is possible by fluorescence in situ hybridization (FISH). A determination is also possible during pregnancy as part of the prenatal diagnosis .
therapy
A causal treatment is not possible due to the genetic cause. However, symptomatic therapy is indicated, in particular the correction of the malformations that occur and the therapy of concomitant diseases.
research
The syndrome was first diagnosed in people with cardiac abnormalities , but was later found in patients with autism and schizophrenia. Scientific research has shown that 20 out of 1000 patients with autism have a 1q21.1 microdeletion.
1q21.1 deletion syndrome is currently being investigated in several locations around the world. There is evidence of a connection between autism and schizophrenia , which could have its origin in embryogenesis through duplication and deletions of chromosomes . Statistical research showed that schizophrenia is significantly more common with a 1q21.1 microdeletion. In contrast, autism is significantly more common with 1q21.1 microduplication. Similar observations were made for chromosome 16 on 16p11.2 (deletion: autism / duplication: schizophrenia) and on chromosome 22 (22q11.21 deletion ( Velo-Cardio-Facial Syndrome ): schizophrenia / duplication: autism) and 22q13.3 (Deletion ( Phelan-McDermid Syndrome ): schizophrenia / duplication: autism). Further research confirms at least a 7.5% probability of an association between schizophrenia and deletions on 1q21.1, 3q29, 15q13.3, 22q11.21 en Neurexin 1 (NRXN1) and duplication on 16p11.2.
Studies on the relationships between autism, schizophrenia and micro changes on chromosome 15 (15q13.3) as well as on chromosome 16 (16p13.1) and chromosome 17 (17p12) are not yet conclusive (status 2011).
Variations in the BCL9 gene, which is located in the distal region, increase the risk of schizophrenia and potentially lead to bipolar disorder and depression.
Research is currently focused on ten to twelve genes on 1q21.1, which are responsible for the production of DUF1220 . DUF1220 is a protein domain of unknown function that is active in the neurons of the brain near the neocortex . Based on research on monkeys and other mammals , it is assumed that the number of DUF1220 gene loci is related to cognitive development (humans: 212; chimpanzees: 37; monkeys: 30; mouse: 1). It appears that the DUF1220 locations on 1q21.1 are in locations related to the size and development of the brain, which in turn is linked to autism ( macrocephaly ) and schizophrenia ( microcephaly ). It is believed that deletion or duplication of a gene that creates DUF1220 areas can cause growth and development disorders in the brain.
In researching the HYDIN2 or HYDIN paralogy , another connection between macrocephaly and duplication and between microcephaly and deletions has been discovered. This part of the 1q21.1 is involved in the development of the brain. It is believed to be a dose-sensitive gene. When this gene is not available in the 1q21.1 range, it leads to microcephaly. The HYDIN2 is a copy of the HYDIN found on 16q22.2.
swell
- Jane Gregory: Bringing up a challenging child at home: when love is not enoug . Jessica Kingsley Publishers, London / Philadelphia, PA 2000, ISBN 1-85302-874-6 .
- Samantha JL Knight (Ed.): Genetics of mental retardation: an overview encompassing learning disability and intellectual disabilit . Karger, Basel / New York 2010, ISBN 978-3-8055-9280-2 .
- HC Mefford, AJ Sharp, C. Baker et al: Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes . In: N. Engl. J. Med. Band 359 , no. October 16 , 2008, p. 1685–1699 , doi : 10.1056 / NEJMoa0805384 , PMID 18784092 , PMC 2703742 (free full text).
- N. Brunetti-Pierri, JS Berg, F. Scaglia et al .: Recurrent reciprocal 1q21.1 deletions and duplications associated with microcephaly or macrocephaly and developmental and behavioral abnormalities . In: Nat. Genet. tape 40 , no. December 12 , 2008, pp. 1466-1471 , doi : 10.1038 / ng.279 , PMID 19029900 , PMC 2680128 (free full text).
- B. Crespi, P. Stead, M. Elliot: Evolution in health and medicine Sackler colloquium: Comparative genomics of autism and schizophrenia . In: Proc. Natl. Acad. Sci. USA . 107 Suppl 1, January 2010, p. 1736–1741 , doi : 10.1073 / pnas.0906080106 , PMID 19955444 , PMC 2868282 (free full text).
- A. Reis, A. Rauch: Chromosomal causes of intellectual disability. In: Medical Genetics. 21 (2009), pp. 237-245, doi: 10.1007 / s11825-009-0166-7
- R. Wimmer, H. Seidel: Chromosomal microdeletions as a cause of pediatric diseases. In: Medical Genetics. 20 (2008), pp. 348-356, doi: 10.1007 / s00112-008-1697-8
Web links
- Chromosome 1q21.1 Deletion Syndrome, 1.35-MB in the OMIM database at Johns Hopkins University
- 1q21.1 recurrent microdeletion (susceptility locus for neurodevelopmental disorders) at DECIPHER
- Brunet et al .: BAC array CGH in patients with Velocardiofacial syndrome-like features reveals genomic aberrations on chromosome region 1q21.1 ; BMC Medical Genetics 2009, 10, p. 144; doi: 10.1186 / 1471-2350-10-144
- Unique: 1q21.1 microdeletions. (PDF; 719 kB) In: www.rarechromo.org. Rare Chromosome Disorder Support Group, 2009, accessed February 2, 2011 .
- GeneReviews NCBI Bookshelf
- Orpha.net
- Blogs about children with 1q21.1 deletion syndrome Chrissy , Laura and Niko
Individual evidence
- ↑ Unique ( Memento of the original from September 2, 2011 in the Internet Archive ) Info: The archive link was inserted automatically and has not yet been checked. Please check the original and archive link according to the instructions and then remove this notice.
- ↑ HC Mefford, AJ Sharp, C. Baker et al.: Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes . In: N Engl J Med . tape 359 , no. October 16 , 2008, p. 1685–1699 , doi : 10.1056 / NEJMoa0805384 , PMID 18784092 , PMC 2703742 (free full text).
- ↑ H. Stefansson, D. Rujescu, S. Cichon et al .: Large recurrent microdeletions associated with schizophrenia . In: Nature . tape 455 , no. 7210 , September 2008, p. 232–236 , doi : 10.1038 / nature07229 , PMID 18668039 , PMC 2687075 (free full text).
- ↑ Jennifer L. Stone, Michael C. O Donovan et al .: Rare chromosomal deletions and duplications increase risk of schizophrenia. In: Nature. 455, 2008, pp. 237-241, doi: 10.1038 / nature07239 .
- ↑ M. Velinov, N. Dolzhanskaya: Clavicular pseudoarthrosis, anomalous coronary artery and extra crease of the fifth finger-previously unreported features in individuals with class II 1q21.1 microdeletions . In: Eur J Med Genet . tape 53 , no. 4 , 2010, p. 213-216 , doi : 10.1016 / j.ejmg.2010.05.005 , PMID 20573555 .
- ↑ SJ Diskin, C. Hou, JT Glessner et al: Copy number variation at 1q21.1 associated with neuroblastoma . In: Nature . tape 459 , no. 7249 , June 2009, p. 987-991 , doi : 10.1038 / nature08035 , PMID 19536264 , PMC 2755253 (free full text).
- ↑ A publication of the Clinic of the Leiden University is expected
- ↑ Lina Basel-Vanagaite et al: An emerging 1q21.1 deletion-associated neurodevelopmental phenotype. In: Journal of Child Neurology. 26 (1), pp. 113-116, doi: 10.1177 / 0883073810377658
- ↑ Douglas F. Levinson et al .: Copy Number Variants in Schizophrenia: Confirmation of Five Previous Findings and New Evidence for 3q29 Microdeletions and VIPR2 Duplications. In: Am J Psychiatry. 2011; 168, pp. 302-316; doi: 10.1176 / appi.ajp.2010.10060876 .
- ↑ Masashi Ikeda et al: Copy Number Variation in Schizophrenia in the Japanese Population. In: Biological Psychiatry. Volume 67, Issue 3, pp. 283-286 (1 February 2010) doi: 10.1016 / j.biopsych.2009.08.034 .
- ↑ Junyan Li et al: Common Variants in the BCL9 Gene Conferring Risk of Schizophrenia. In: Arch Gen Psychiatry . 2011; 68 (3), pp. 232-240. doi: 10.1001 / archgenpsychiatry.2011.1 .