Thermolysin
| Thermolysin | ||
|---|---|---|

|
||
| Belt model according to PDB 2A7G . The zinc binding sites are yellow, other catalytic amino acids orange, zinc light blue, calcium salmon-colored. | ||
|
Existing structural data: see UniProt entry |
||
| Mass / length primary structure | 316 amino acids | |
| Cofactor | Zn 2+ ; 4 Ca 2+ | |
| Identifier | ||
| Gene name (s) | npr | |
| External IDs | ||
| Enzyme classification | ||
| EC, category | 3.4.24.27 , metalloprotease | |
| MEROPS | M04.001 | |
| Substrate | Xaa - + - Leu> Xaa - + - Phe | |
| Occurrence | ||
| Homology family | Thermolysin | |
| Parent taxon | bacteria | |
Thermolysin is a thermostable enzyme that belongs to the group of metallopeptidases . It is secreted by the thermophilic bacterium Bacillus thermoproteolyticus . As an endoprotease , it catalyzes the cleavage of peptide bonds ; The hydrolysis of the peptide bond takes place before large, hydrophobic residues (P1 'activity) (for example after leucine , isoleucine or phenylalanine ) within the polypeptide.
The enzyme has the EC number EC 3.4.24.27 .
General
Thermolysin belongs to the zinc- containing metalloproteases and is one of the best-studied enzymes in this superfamily. The three-dimensional structure was already clarified in 1972 by Matthews and co-workers in a resolution of 2.3 Å , meanwhile a resolution of 1.6 Å has been achieved.
structure
Thermolysin has a molar mass of 34.6 kDa . The enzyme has an N-terminal and a C-terminal domain that form a column for its active center. The N-terminal domain consists mainly of β-sheets , the more stable C-terminal domain of α-helices .
Thermolysin requires one zinc ion and four calcium ions . The former is essential for enzyme activity, but hardly contributes to thermal stability . The zinc ion is found in the active center. The thermal stability is mediated by the calcium ions. The calcium ions stabilize the structural integrity and protect the loops protruding from the surface of the enzyme from autoproteolysis ("self-digestion").
The protein has no disulfide bridges.
Chemical properties
The endoprotease is thermally stable, its function is optimal between 65 and 70 ° C at pH 8.0. At 80 ° C, its structural half-life is one hour. However, a heat denaturation of thermolysin takes place quickly and irreversibly. It does this through unfolding and autoproteolysis of exposed sequences.
Catalysis mechanism
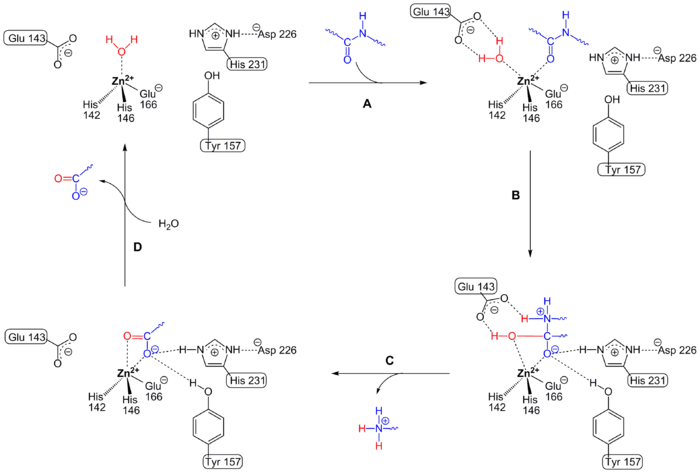
Although there is a lot of data on crystal structure analyzes , mutagenesis experiments and investigations of the reaction kinetics, the exact catalytic mechanism is still open to debate. With the help of computer simulations from crystallographic data, however, the following reaction mechanism is considered to be the most likely.
In the native state, the zinc ion is tetrahedrally coordinated . The metal ion forms a complex with three amino acid residues of the enzyme ( histidine 142 (His 142), histidine 146 (His 146) and glutamic acid 166 (Glu 166)) and a water molecule (see also the figure below). The latter is common to all zinc metalloproteins.
After binding the peptide to be hydrolyzed (step 1 in the figure), the zinc ion is pentacoordinated. At the same time, the water molecule is brought close to the glutamate 143 (Glu 143) and is strongly polarized by this and the positively charged zinc atom. As a result, the nucleophilicity of the water molecule becomes large enough to attack the peptide bond.
The nucleophilic attack (step 2 ) on the peptide bond takes place, during which one proton of the water is transferred to the nitrogen by the Glu 143. The carbon atom of the peptide bond is now tetrahedral. His 231 and tyrosine (Tyr) 157 help stabilize carbonyl oxygen. Finally, the peptide bond is cleaved, and the second proton from the water is transferred to the nitrogen atom via Glu 143 .
The amine product leaves the active site as a protonated form (step 3 ). The carboxyl product is replaced by a new water molecule (step 4 ), so that the zinc atom is again tetrahedrally coordinated. A new reaction mechanism can begin.
The Glu-143 residue forms a so-called matching sequence motif with the two histidines His 142 and 146, the HE XX H motif . This motif is highly conserved in all representatives of zinc endoproteases with a zinc atom.
Inhibitors
A natural inhibitor of thermolysin is phosphoramidon from Streptomyces tanashiensis . The currently most effective inhibitor is the artificial ZF p LA (Carbobenzoxy- L -Phe P - L -Leu- L -Ala). In both cases there is a phosphoramidate (a covalent bond between a nitrogen and a phosphorus atom).
Technical applications
The enzyme can be used for the synthesis of aspartame , a sweetener. The condensation - i.e. the reverse reaction of hydrolysis ( reverse proteolysis ) - of L- aspartate and L -phenylalanine to L -aspartyl- L -phenylalanine methyl ester (= aspartame) is catalyzed.
literature
- Matthews, BW. (1988): Structural basis of the action of thermolysin and related zinc peptidases . In: Acc. Chem. Res. 21 (9); 333-340; doi: 10.1021 / ar00153a003
- Pelmenschikov, V. et al. (2002): A theoretical study of the mechanism for peptide hydrolysis by thermolysin . J Biol Inorg Chem. 7 (3); 284-98; PMID 11935352 ; doi: 10.1007 / s007750100295
Web links
Individual evidence
- ↑ Endo, S. (1962). Studies on protease produced by thermophilic bacteria . In: J. Ferment. Technol. 40 ; 346-353.
- ↑ Morihara, K. and Tsuzuki, H. (1970): Thermolysin: kinetic study with oligopeptides . In: Eur J Biochem . 15 (2); 374-80; PMID 4993757 .
- ^ Entry of thermolysin on ExPASy
- ^ Colman PM. et al. (1972): The structure of thermolysin: an electron density map at 2-3 Å resolution . In: J Mol Biol. 70 (3): 701-24; PMID 5083153 .
- ^ Weaver LH. et al. (1976): The structure and stability of thermolysin . In: Experientia Suppl. 26 ; 31-9; PMID 820566 .
- ↑ a b Roche, RS. and Voordouw, G. (1978): The structural and functional roles of metal ions in thermolysin . In: CRC Crit Rev Biochem. 5 (1); 1-23; PMID 357082 .
- ↑ Tajima, M. et al. (1976): Role of calcium ions in the thermostability of thermolysin and Bacillus subtilis var. Amylosacchariticus neutral protease . In: Eur J Biochem. 64 (1); 243-7; PMID 819262 .
- ↑ W. Kaim / B. Schwederski, Bioinorganische Chemie, Teubner Study Books, 1995, ISBN 3-519-13505-1 , p. 261.
- ↑ Thermolysin data sheet ( Memento from February 19, 2014 in the Internet Archive ) (PDF; 104 kB) from Daiwa Kasei KK
- ↑ a b Rao, MB. et al. (1998): Molecular and biotechnological aspects of microbial proteases . In: Microbiol Mol Biol Rev. 62 (3); 597-635; PMID 9729602 ; PDF (free full text access).
- ↑ External identifiers of or database links for phosphoramidon : CAS number: 36357-77-4, EC number: 252-996-3, ECHA InfoCard: 100.048.164 , PubChem : 445114 , ChemSpider : 392848 , DrugBank : DB02557 , Wikidata : Q7187544 .
- ↑ External identifiers of or database links to ZF p LA, also 4tmn : CAS number: 110786-00-0, PubChem : 5492587 , ChemSpider : 4591060 , Wikidata : Q27451311 .
- ↑ Arnold Ulrich: Limited proteolysis for the analysis of local and global changes in the protein structure using the example of ribonucleases. Martin Luther University Halle-Wittenberg, habilitation thesis, 2008, pp. 35–36.