Transcription (biology)
| Parent |
|
Gene expression RNA biosynthesis |
| Subordinate |
|
Initiation Elongation Termination |
| Gene Ontology |
|---|
| QuickGO |
In genetics, the synthesis of RNA based on DNA as a template is called transcription (from late Latin transcriptio "transmission" to Latin transcribere "rewrite / overwrite") . The resulting RNA can largely be divided into three groups: mRNA (messenger RNA) (for protein biosynthesis ) as well as tRNA (transfer RNA) and rRNA (ribosomal RNA). Like translation , transcription is an essential part of gene expression .

In transcription is a gene read and reproduced as an RNA molecule, that is a specific DNA segment serves as a template ( template , English template ) for the synthesis of a new strand of RNA. During this process, the nucleobases of DNA (A - T - G - C) are transcribed into the nucleobases of RNA (U - A - C - G). Instead of thymine is uracil and in place of the deoxyribose is ribose in RNA before.
The process of transcription is basically the same for eukaryotes and prokaryotes . There are differences in the control and in the subsequent modification. In the case of prokaryotes, the control takes place via an operator , while in the case of eukaryotes the regulation can be regulated via an enhancer or silencer , which is upstream or downstream of the promoter . Furthermore, in prokaryotes, transcription takes place in the cytoplasm of the cell, in eukaryotes in the cell nucleus ( karyoplasm ). In eukaryotes, the pre-mRNA is also processed during or after its synthesis before it is transported from the cell nucleus into the cytoplasm. After transcription, the mRNA is translated into a protein in the cytoplasm on the ribosome .
Synthesis of the mRNA
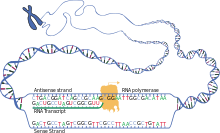
- Synthesis of the mRNA:
- General procedure: The RNA polymerase - protein complex consists of a promoter, said DNA region (see the main article Initiation ). The RNA polymerase uncoils the double helix and thus exposes approx. 10–20 bases of the DNA for pairing. On the codogenic strand (Fig .: antisense strand) of the DNA, complementary ribonucleotides are attached by base pairing . With the elimination of pyrophosphate from the nucleoside triphosphates, they are linked to one another by an ester bond between phosphate and ribose. The reading direction of the DNA runs from the 3 'end to the 5' end , the synthesis of the complementary RNA accordingly from 5 'to 3' (see main article elongation ). The RNA polymerase does not require a primer, the transcription is terminated at the terminator. The mRNA transcript is then released and the polymerase detaches from the DNA (see the main article Termination ).
- In the case of eukaryotic mRNA synthesis, in addition to the process just described, a cap structure at the 5 'end of the mRNA is synthesized , which protects it and acts as a signal for export from the cell nucleus. This so-called capping already happens when the transcript is only a few nucleotides long, i.e. before elongation.
- Further processing of the mRNA:
- In prokaryotes, the mRNA reaches the ribosomes directly after the copying process, and ribosomes are often already attached to the chain that is still being formed and translation begins before the transcription is complete (poly-ribosome complex).
- In eukaryotes, the RNA transcript itself does not yet leave the nucleus. The RNA produced in the first part of the transcription is called immature RNA, pre-mRNA or hnRNA (heterogeneous nuclear RNA). It is still processed at the 3 'end by polyadenylation (tailing) and splicing . By alternative splicing different mRNA molecules may arise from the same DNA segment. The so-called mature mRNA leaves the cell nucleus through a nuclear pore and thus reaches the cytoplasm , where it can interact with the ribosomes.
Synthesis of the tRNA and the rRNA
Transfer RNA (tRNA) and ribosomal RNA (rRNA) are synthesized on the DNA by two other RNA polymerases, both of which work on a different principle than that of RNA polymerase II . They resemble those of the prokaryotes, in which the same RNA polymerase acts as a catalyst . In eukaryotes, the synthesis of the tRNA, the 5S rRNA and the 7SL-RNA is carried out by the RNA polymerase III, the synthesis of the rRNA and partly also the sn-RNA (small nuclear RNA) by the RNA polymerase I, the synthesis of the m-RNA by RNA polymerase II.
End of transcription
In eukaryotic cells, the RNA polymerase cannot recognize the end of a gene by itself; it needs auxiliary factors that interact with the polymerase. These protein complexes recognize the polyadenylation site ( 5'-AAUAAA-3 ' ), cut the RNA and initiate polyadenylation while the RNA polymerase continues to work at the same time. A model for the termination of transcription is that the still growing, useless end of the RNA is degraded by an exonuclease ( Rat1 ) faster than it is elongated by the polymerase. When the exonuclease reaches the transcription site, the polymerase detaches itself from the DNA, and transcription is finally terminated (torpedo model of transcriptional termination). In addition, other protein complexes (e.g. TREX) seem to be important for a termination.
Archaeal transcription
The archaea genes have a consensus sequence called a TATA box in the promoter . The two initiation factors of the archaea, TBP and TFB, bind to the promoter. A polymerase, which is orthologous to the eukaryotic RNA polymerase II and consists of twelve subunits, binds to this in turn.
In many cases, the transcription is linked to the subsequent splicing of the mRNA before the mRNA swims out of a nuclear pore or is now directly further synthesized into a protein.
Bacterial transcription
In contrast to the eukaryotes, bacteria only have one RNA polymerase. The core or minimal enzyme consists of four subunits (2 × α, β, β '), which catalyzes transcription but does not initiate it. The core of the enzyme acts reciprocally with the loose sigma subunit and the holo-enzyme (2 × α, β, β ', σ (Sigma)) is formed, which can carry out the initiation (Sigma allows gliding along the DNA and Finding the Promoter's Pribnow Box ). The sigma subunit binds to the promoter of the non-template strand and there breaks the hydrogen bonds between the base pairs; it has a helicase function, which is the most important function of this polymerase.
The function of the α-subunit is, on the one hand, the preservation and stability of the structure, due to the amino terminal domain, and, on the other hand, the carboxy-terminal domain binds to the promoter and interacts with transcription regulatory elements.
The β and β 'subunits work together to provide binding to the DNA template and a growing RNA chain.
The σ (sigma) subunit recognizes transcription origins. There are several sigma subunits that recognize different groups of genes. The σ 70 subunit is the most widespread .
The ω (omega) subunit serves to stabilize and maintain the structure and is not absolutely necessary.
Reverse transcription
In some viruses there is the special case of reverse transcription, in which DNA is produced from an RNA template.
literature
- Rolf Knippers: Molecular Genetics. 9th completely revised edition. Thieme, Stuttgart et al. 2006, ISBN 3-13-477009-1 , pp. 49-58.
Web links
- W. H. Freeman: Animation of the Transcription , www.sumanasinc.com
- Headquarters for teaching media on the Internet e. V. (ZUM) : Transcription / Genetic Code , www.zum.de