Polymerase chain reaction: Difference between revisions
→PCR in practice: reworded |
No edit summary |
||
| Line 1: | Line 1: | ||
'''Bold text'''''Italic text''[[Link title]][[Link title]]{{redirect|PCR}}[http://www.example.com link title][[Link title]]''Italic text'' |
|||
{{redirect|PCR}} |
|||
'''Polymerase chain reaction''' ('''PCR''') is a [[molecular biology]] technique <ref>[http://www.si.edu/archives/ihd/videocatalog/9577.htm The history of PCR]: [[Smithsonian Institution]] Archives, Institutional History Division. Retrieved 24 June 2006.</ref> for [[enzyme|enzymatically]] [[DNA replication|replicating]] [[DNA]] without using a living [[organism]], such as ''[[E. coli]]'' or [[yeast]]. Like amplification using living organisms, the technique allows a small amount of DNA to be amplified exponentially. As PCR is an ''[[in vitro]]'' technique, it can be performed without restrictions on the form of DNA and it can be extensively modified to perform a wide array of [[Genetic engineering|genetic manipulations]]. |
'''Polymerase chain reaction''' ('''PCR''') is a [[molecular biology]] technique <ref>[http://www.si.edu/archives/ihd/videocatalog/9577.htm The history of PCR]: [[Smithsonian Institution]] Archives, Institutional History Division. Retrieved 24 June 2006.</ref> for [[enzyme|enzymatically]] [[DNA replication|replicating]] [[DNA]] without using a living [[organism]], such as ''[[E. coli]]'' or [[yeast]]. Like amplification using living organisms, the technique allows a small amount of DNA to be amplified exponentially. As PCR is an ''[[in vitro]]'' technique, it can be performed without restrictions on the form of DNA and it can be extensively modified to perform a wide array of [[Genetic engineering|genetic manipulations]]. |
||
Revision as of 23:45, 6 November 2006
Bold textItalic textLink titleLink title
link titleLink titleItalic text
Polymerase chain reaction (PCR) is a molecular biology technique [1] for enzymatically replicating DNA without using a living organism, such as E. coli or yeast. Like amplification using living organisms, the technique allows a small amount of DNA to be amplified exponentially. As PCR is an in vitro technique, it can be performed without restrictions on the form of DNA and it can be extensively modified to perform a wide array of genetic manipulations.
PCR is commonly used in medical and biological research labs for a variety of tasks, such as the detection of hereditary diseases, the identification of genetic fingerprints, the diagnosis of infectious diseases, the cloning of genes, paternity testing, and DNA computing.
PCR was invented by Kary Mullis. At the time he thought up PCR in 1983, Mullis was working in Emeryville, California for Cetus, one of the first biotechnology companies. There, he was charged with making short chains of DNA for other scientists. Mullis has written that he conceived of PCR while cruising along the Pacific Coast Highway 1 one night in his car. He was playing in his mind with a new way of analyzing changes (mutations) in DNA when he realized that he had instead invented a method of amplifying any DNA region. Mullis has said that before his trip was over, he was already savoring the prospects of a Nobel Prize. He shared the Nobel Prize in Chemistry with Michael Smith in 1993.
As Mullis has written in the Scientific American: "Beginning with a single molecule of the genetic material DNA, the PCR can generate 100 billion similar molecules in an afternoon. The reaction is easy to execute. It requires no more than a test tube, a few simple reagents, and a source of heat."
PCR in practice
PCR is used to amplify specific regions of a DNA strand. This can be a single gene, just a part of a gene, or non-coding sequence. PCR processes usually amplifies only short DNA fragments, usually up to 10 kb. Certain methods can copy fragments up to 47 kb in size[citation needed], which is still much less than the chromosomal DNA of a eukaryotic cell - for example, a human cell contains about three billion base pairs.
PCR, as currently practiced, requires several basic components. These components are:
- DNA template, which contains the region of the DNA fragment to be amplified
- Two primers, which determine the beginning and end of the region to be amplified (see following section on primers)
- Taq polymerase (or another durable polymerase), a DNA polymerase, which copies the region to be amplified
- Deoxynucleotides-triphosphate, from which the DNA Polymerase builds the new DNA
- Buffer, which provides a suitable chemical environment for the DNA Polymerase
The PCR process is carried out in a thermal cycler. This is a machine that heats and cools the reaction tubes within it to the precise temperature required for each step of the reaction. To prevent evaporation of the reaction mixture (typically volumes between 15-100µl per tube), a heated lid is placed on top of the reaction tubes or a layer of oil is put on the surface of the reaction mixture. These machines cost more than $2,500 USD, as of 2004.
Primers
The DNA fragment to be amplified is determined by selecting primers. Primers are short, artificial DNA strands — often not more than 50 and usually only 18 to 25 base pairs long — that are complementary to the beginning or the end of the DNA fragment to be amplified. They anneal by adhering to the DNA template at these starting and ending points, where the DNA polymerase binds and begins the synthesis of the new DNA strand.
The choice of the length of the primers and their melting temperature (Tm) depends on a number of considerations. The melting temperature of a primer -- not to be confused with the melting temperature of the template DNA -- is defined as the temperature at which half of the primer binding sites are occupied. Primers that are too short would anneal at several positions on a long DNA template, which would result in non-specific copies. On the other hand, the length of a primer is limited by the maximum temperature allowed to be applied in order to melt it, as melting temperature increases with the length of the primer. Melting temperatures that are too high, i.e., above 80°C, can cause problems since the DNA polymerase is less active at such temperatures. The optimum length of a primer is generally from 15 to 40 nucleotides with a melting temperature between 55°C and 65°C.
Sometimes degenerate primers are used. These are actually mixtures of similar, but not identical, primers. They may be convenient if the same gene is to be amplified from different organisms, as the genes themselves are probably similar but not identical. The other use for degenerate primers is when primer design is based on protein sequence. As several different codons can code for one amino acid, it is often difficult to deduce which codon is used in a particular case. Therefore primer sequence corresponding to the amino acid isoleucine might be "ATH", where A stands for adenine, T for thymine, and H for adenine, thymine, or cytosine. (See genetic code for further details about codons.) Use of degenerate primers can greatly reduce the specificity of the PCR amplification. This problem can be partly solved by using touchdown PCR.
The above mentioned considerations make primer design a very exacting process, upon which product yield depends:
- GC-content should be between 40-60%.
- Calculated Tm for both primers used in reaction should not differ >5°C and Tm of the amplification product should not differ from primers by >10°C.
- Annealing temperature usually is 5°C below the calculated lower Tm. However it should be chosen empirically for individual conditions.
- Inner self-complementary hairpins of >4 and of dimers >8 should be avoided.
- Primer 3' terminus design is critical to PCR success since the primer extends from the 3' end. The 3' end should not be complementary over greater than 3-4 bases to any region of the other primer (or even the same primer) used in the reaction and must provide correct base matching to template.
There are computer programs to help design primers (see External links).
Procedure
The PCR process usually consists of a series of twenty to thirty-five cycles. Each cycle consists of three steps (Fig. 2).
- The double-stranded DNA has to be heated to 94-96°C (or 98°C if extremely thermostable polymerases are used) in order to separate the strands. This step is called denaturing; it breaks apart the hydrogen bonds that connect the two DNA strands. Prior to the first cycle, the DNA is often denatured for an extended time to ensure that both the template DNA and the primers have completely separated and are now single-strand only. Time: usually 1-2 minutes, but up to 5 minutes. Also certain polymerases are activated at this step (see hot-start PCR).
- After separating the DNA strands, the temperature is lowered so the primers can attach themselves to the single DNA strands. This step is called annealing. The temperature of this stage depends on the primers and is usually 5°C below their melting temperature (45-60°C). A wrong temperature during the annealing step can result in primers not binding to the template DNA at all, or binding at random. Time: 1-2 minutes.
- Finally, the DNA polymerase has to copy the DNA strands. It starts at the annealed primer and works its way along the DNA strand. This step is called elongation. The elongation temperature depends on the DNA polymerase. Taq polymerase elongates optimally at a temperature of 72 Celsius. The time for this step depends both on the DNA polymerase itself and on the length of the DNA fragment to be amplified. As a rule-of-thumb, this step takes 1 minute per thousand base pairs. A final elongation step is frequently used after the last cycle to ensure that any remaining single stranded DNA is completely copied. This differs from all other elongation steps, only in that it is longer, typically 10-15 minutes. This last step is highly recomendable if the PCR product is to be ligated into a T vector using TA-cloning.

Example
The times and temperatures given in this example are taken from a PCR program that was successfully used on a 250 bp fragment of the C-terminus of the insulin-like growth factor (IGF)[citation needed].
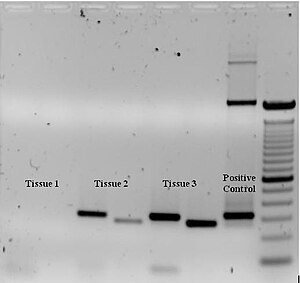
The reaction mixture consists of
- 1.0 µl DNA template (100 ng/µl)
- 2.5 µl of primer, 1.25 µl per primer (100 ng/µl)
- 1.0 µl Pfu-Polymerase
- 1.0 µl nucleotides
- 5.0 µl buffer solution
- 89.5 µl water
A 200 µl reaction tube containing the 100 µl mixture is inserted into the thermocycler.
The PCR process consists of the following steps:
- Initialization. The mixture is heated at 96°C for 5 minutes to ensure that the DNA strands as well as the primers have melted. The DNA Polymerase can be present at initialization, or it can be added after this step.
- Melting, where it is heated at 96°C for 30 seconds. For each cycle, this is usually enough time for the DNA to denature.
- Annealing by heating at 68°C for 30 seconds:The primers are jiggling around, caused by the Brownian motion. Short bondings are constantly formed and broken between the single stranded primer and the single stranded template. The more stable bonds last a little bit longer (primers that fit exactly) and on that little piece of double stranded DNA (template and primer), the polymerase can attach and starts copying the template. Once there are a few bases built in, the Tm of the double-stranded region between the template and the primer is greater than the annealing or extension temperature.
- Elongation by heating 72°C for 45 seconds:This is the ideal working temperature for the polymerase. The primers, having been extended for a few bases, already have a stronger hydrogen bond to the template than the forces breaking these attractions. Primers that are on positions with no exact match, melt away from the template (because of the higher temperature) and are not extended.
The bases (complementary to the template) are coupled to the primer on the 3' side (the polymerase adds dNTP's from 5' to 3', reading the template from 3' to 5' side, bases are added complementary to the template)
- Steps 2-4 are repeated 25 times, but with good primers and fresh polymerase, 15 to 20 cycles is sufficient.
- Mixture is held at 7°C. This is useful if one starts the PCR in the evening just before leaving the lab, so it can run overnight. The DNA will not be damaged at 7°C after just one night.
The PCR product can be identified by its size using agarose gel electrophoresis. Agarose gel electrophoresis is a procedure that consists of injecting DNA into agarose gel and then applying an electric current to the gel. As a result, the smaller DNA strands move faster than the larger strands through the gel toward the positive current. The size of the PCR product can be determined by comparing it with a DNA ladder, which contains DNA fragments of known size, also within the gel (Fig. 3).
PCR optimization
Since PCR is very sensitive, adequate measures to avoid contamination from other DNA present in the lab environment (bacteria, viruses, lab staff's skin etc.) should be taken. Thus DNA sample preparation, reaction mixture assemblage and the PCR process, in addition to the subsequent reaction product analysis, should be performed in separate areas. For the preparation of reaction mixture, a laminar flow cabinet with UV lamp is recommended. Fresh gloves should be used for each PCR step as well as displacement pipettes with aerosol filters. The reagents for PCR should be prepared separately and used solely for this purpose. Aliquots should be stored separately from other DNA samples. A control reaction (inner control), omitting template DNA, should always be performed, to confirm the absence of contamination or primer multimer formation.
Difficulties with polymerase chain reaction
Polymerase chain reaction is not perfect, and errors and mistakes can occur. These are some common errors and problems that may occur.
Polymerase errors
Taq polymerase lacks a 3' to 5' exonuclease activity. This makes it impossible for it to check the base it has inserted and remove it if it is incorrect, a process common in higher organisms. This in turn results in a high error rate of approximately 1 in 10,000 bases, which, if an error occurs early, can alter large proportions of the final product.
Other polymerases are available for accuracy in vital uses such as amplification for sequencing. Examples of polymerases with 3'to 5' exonuclease activity include: KOD DNA polymerase, a recombinant form of Thermococcus kodakaraensis KOD1; Vent, which is extracted from Thermococcus litoralis; Pfu DNA polymerase, which is extracted from Pyrococcus furiosus; and Pwo, which is extracted from Pyrococcus woesii.
Size limitations
PCR works readily with DNA of lengths two to three thousand basepairs, but above this length the polymerase tends to fall off and the typical heating cycle does not leave enough time for polymerisation to complete. It is possible to amplify larger pieces of up to 50,000 base pairs, with a slower heating cycle and special polymerases. These special polymerases are often polymerases fused to a DNA-binding protein, making them literally "stick" to the DNA longer.
Non specific priming
The non specific binding of primers is always a possibility due to sequence duplications, non-specific binding and partial primer binding, leaving the 5' end unattached. This is increased by the use of degenerate sequences or bases in the primer. Manipulation of annealing temperature and magnesium ion (which stabilise DNA and RNA interactions) concentrations can increase specificity. Non-specific priming can be prevented during the low temperatures of reaction preparation by use of "hot-start" polymerase enzymes where the active site is blocked by an antibody or chemical that only dislodges once the reaction is heated to 95˚C during the denaturation step of the first cycle.
Other methods to increase specificity include Nested PCR and Touchdown PCR.
Practical modifications to the PCR technique
- Nested PCR - Nested PCR is intended to reduce the contaminations in products due to the amplification of unexpected primer binding sites. Two sets of primers are used in two successive PCR runs, the second set intended to amplify a secondary target within the first run product. This is very successful, but requires more detailed knowledge of the sequences involved.
- Ligation-mediated PCR
- Inverse PCR - Inverse PCR is a method used to allow PCR when only one internal sequence is known. This is especially useful in identifying flanking sequences to various genomic inserts. This involves a series of digestions and self ligation before cutting by an endonuclease, resulting in known sequences at either end of the unknown sequence.
- RT-PCR - RT-PCR (Reverse Transcription PCR) is the method used to amplify, isolate or identify a known sequence from a cell or tissues RNA library. Essentially normal PCR preceded by transcription by Reverse transcriptase (to convert the RNA to cDNA) this is widely used in expression mapping, determining when and where certain genes are expressed.
- Assembly PCR - Assembly PCR is the completely artificial synthesis of long gene products by performing PCR on a pool of long oligonucleotides with short overlapping segments. The oligonucleotides alternate between sense and antisense directions, and the overlapping segments serve to order the PCR fragments so that they selectively produce their final product.
- Asymmetric PCR - Asymmetric PCR is used to preferentially amplify one strand of the original DNA more than the other. It finds use in some types of sequencing and hybridization probing where having only one of the two complementary stands is ideal. PCR is carried out as usual, but with a great excess of the primers for the chosen strand. Due to the slow (arithmetic) amplification later in the reaction after the limiting primer has been used up, extra cycles of PCR are required. A recent modification on this process, known as Linear-After-The-Exponential-PCR (LATE-PCR), uses a limiting primer with a higher melting temperature (Tm) than the excess primer to maintain reaction efficiency as the limiting primer concentration decreases mid-reaction.
- Quantitative PCR - Q-PCR (Quantitative PCR) is used to rapidly measure the quantity of PCR product (preferably real-time), thus is an indirect method for quantitatively measuring starting amounts of DNA, cDNA or RNA. This is commonly used for the purpose of determining whether a sequence is present or not, and if it is present the number of copies in the sample. There are 3 main methods which vary in difficulty and detail.
- Quantitative real-time PCR is often confusingly known as RT-PCR (Real Time PCR) and RQ-PCR. QRT-PCR or RTQ-PCR are more appropriate contractions. RT-PCR can also refer to reverse transcription PCR, which even more confusingly, is often used in conjunction with Q-PCR. This method uses fluorescent dyes and probes to measure the amount of amplified product in real time.
- Touchdown PCR - Touchdown PCR is a variant of PCR that reduces nonspecific primer annealing by more gradually lowering the annealing temperature between cycles. As higher temperatures give greater specificity for primer binding, primers anneal first as the temperature passes through the zone of greatest specificity.
- Hot-start PCR is a technique that reduces non-specific priming that occurs during the preparation of the reaction components. The technique may be performed manually by simply heating the reaction components briefly at the melting temperature before adding the polymerase. Specialized enzyme systems have been developed that inhibit the polymerase's activity at ambient temperature, either by the binding of an antibody or by the presence of covalently bound inhibitors that only dissociate after a high-temperature activation step.
- Colony PCR - Bacterial clones (E.coli) can be screened for the correct ligation products. Selected colonies are picked with a sterile toothpick from an agarose plate and dabbed into the master mix or sterile water. Primers (and the master mix) are added - the PCR protocol has to be started with an extended time at 95^^C.
- RACE-PCR - Rapid amplification of cDNA ends.
- Multiplex-PCR - The use of multiple, unique primer sets within a single PCR reaction to produce amplicons of varying sizes specific to different DNA sequences. By targeting multiple genes at once, additional information may be illicited from a single test run that otherwise would require several times the reagents and technician time to perform. Annealing temperatures for each of the primer sets must be optimized to work correctly within a single reaction and amplicon sizes should be separated by enough difference in final base pair length to form distinct bands via gel electrophoresis.
- Methylation Specific PCR - Methylation Specific PCR (MSP) is used to detect methylation of CpG islands in genomic DNA. DNA is first treated with sodium bisulfite, which converts unmethylated cytosine bases to uracil, which is recognized by PCR primers as thymine. Two PCR reactions are then carried out on the modified DNA, using primer sets identical except at any CpG islands within the primer sequences. At these points, one primer set recognizes DNA with cytosines to amplify methylated DNA, and one set recognizes DNA with uracil or thymine to amplify unmethylated DNA. MSP using qPCR can also be performed to obtain quantitative rather than qualitative information about methylation.
Recent developments in PCR techniques
- A more recent method which excludes a temperature cycle, but uses enzymes, is helicase-dependent amplification.
- TAIL-PCR, developed by Liu et al. in 1995, is the thermal asymmetric interlaced PCR.
- Meta-PCR, developed by Andrew Wallace, allows to optimize amplification and direct sequence analysis of complex genes. Details at National Genetic Reference Laboratory, Manchester, UK
Uses of PCR
PCR can be used for a broad variety of experiments and analyses. Some examples are discussed below.
Genetic fingerprinting
Genetic fingerprinting is a forensic technique used to identify a person by comparing his or her DNA with a given sample, such as blood from a crime scene can be genetically compared to blood from a suspect. The sample may contain only a tiny amount of DNA, obtained from a source such as blood, semen, saliva, hair, or other organic material. Theoretically, just a single strand is needed. First, one breaks the DNA sample into fragments, then amplifies them using PCR. The amplified fragments are then separated using gel electrophoresis. The overall layout of the DNA fragments is called a DNA fingerprint. Since there is a very small possibility that two individuals may have the same sequences, the technique is more effective at acquitting a suspect than proving the suspect guilty. This small possibility was exploited by defense lawyers in the controversial O.J. Simpson case. A match however usually remains a very strong indicator also in the question of guilt.
Paternity testing

Although these resulting 'fingerprints' are unique (except for identical twins), genetic relationships, for example, parent-child or siblings, can be determined from two or more genetic fingerprints, which can be used for paternity tests (Fig. 4). A variation of this technique can also be used to determine evolutionary relationships between organisms.
Detection of hereditary diseases
The detection of hereditary diseases in a given genome is a long and difficult process, which can be shortened significantly by using PCR. Each gene in question can easily be amplified through PCR by using the appropriate primers and then sequenced to detect mutations.
Viral diseases, too, can be detected using PCR through amplification of the viral DNA. This analysis is possible right after infection, which can be from several days to several months before actual symptoms occur. Such early diagnoses give physicians a significant lead in treatment.
Cloning genes
Cloning a gene, not to be confused with cloning a whole organism, describes the process of isolating a gene from one organism and then inserting it into another organism (now termed a genetically modified organism (GMO)). PCR is often used to amplify the gene, which can then be inserted into a vector (a vector is a piece of DNA which 'carries' the gene into the GMO) such as a plasmid (a circular DNA molecule) (Fig. 5). The DNA can then be transferred into an organism (the GMO) where the gene and its product can be studied more closely. Expressing a cloned gene (when a gene is expressed the gene product (usually protein or RNA) is produced by the GMO) can also be a way of mass-producing useful proteins, for example medicines or the enzymes in biological washing powders. The incorporation of an affinity tag on a recombinant protein will generate a fusion protein which can be more easily purified by affinity chromatography.

(1) Chromosomal DNA of organism A. (2) PCR. (3) Multiple copies of a single gene from organism A. (4) Insertion of the gene into a plasmid. (5) Plasmid with gene from organism A. (6) Insertion of the plasmid in organism B. (7) Multiplication or expression of the gene, originally from organism A, occurring in organism B.
Mutagenesis
Mutagenesis is a way of making changes to the sequence of nucleotides in the DNA. There are situations in which one is interested in mutated (changed) copies of a given DNA strand, for example, when trying to assess the function of a gene or in in-vitro protein evolution (also known as Directed evolution). Mutations can be introduced into copied DNA sequences in two fundamentally different ways in the PCR process. Site-directed mutagenesis allows the experimenter to introduce a mutation at a specific location on the DNA strand. Usually, the desired mutation is incorporated in the primers used for the PCR program. Random mutagenesis, on the other hand, is based on the use of error-prone polymerases in the PCR process. In the case of random mutagenesis, the location and nature of the mutations cannot be controlled. One application of random mutagenesis is to analyze structure-function relationships of a protein. By randomly altering a DNA sequence, one can compare the resulting protein with the original and determine the function of each part of the protein.
Analysis of ancient DNA
Using PCR, it becomes possible to analyze DNA that is thousands of years old. PCR techniques have been successfully used on animals, such as a forty-thousand-year-old mammoth, and also on human DNA, in applications ranging from the analysis of Egyptian mummies to the identification of a Russian Tsar.
Genotyping of specific mutations
Through the use of allele-specific PCR, one can easily determine which allele of a mutation or polymorphism an individual has. Here, one of the two primers is common, and would anneal a short distance away from the mutation, while the other anneals right on the variation. The 3' end of the allele-specific primer is modified, to only anneal if it matches one of the alleles. If the mutation of interest is a T or C single nucleotide polymorphism (T/C SNP), one would use two reactions, one containing a primer ending in T, and the other ending in C. The common primer would be the same. Following PCR, these two sets of reactions would be run out on an agarose gel, and the band pattern will tell you if the individual is homozygous T, homozygous C, or heterozygous T/C. This methodology has several applications, such as amplifying certain haplotypes (when certain alleles at 2 or more SNPs occur together on the same chromosome Linkage Disequilibrium) or detection of recombinant chromosomes and the study of meiotic recombination.
Comparison of gene expression
Researchers have used traditional PCR as a way to estimate changes in the amount of a gene's expression. Ribonucleic acid (RNA) is the molecule into which DNA is transcribed prior to making a protein, and those strands of RNA that hold the instructions for protein sequence are known as messenger RNA (mRNA). Once RNA is isolated it can be reverse transcribed back into DNA (complementary DNA to be precise, known as cDNA), at which point traditional PCR can be applied to amplify the gene, this methodology is called RT-PCR. In most cases if there is more starting material (mRNA) of a gene then during PCR more copies of the gene will be generated. When the products of the PCR process are run on an agarose gel (see Figure 3 above) a band, corresponding to a gene, will appear larger on the gel (note that the band remains in the same location relative to the ladder, it will just appear fatter or brighter). By running samples of amplified cDNA from differently treated organisms one can get a general idea of which sample expressed more of the gene of interest. A quantative RT-PCR method has been developed, it is called Real-time PCR .
History
Polymerase chain reaction was invented by Kary Mullis. He was awarded the Nobel Prize in Chemistry in 1993 for his invention, only seven years after he and his colleagues at Cetus first reduced his proposal to practice. The idea was to develop a process by which DNA could be artificially multiplied through repeated cycles of duplication driven by an enzyme called DNA polymerase.
DNA polymerase occurs naturally in living organisms. In cells it functions to duplicate DNA when cells divide in mitosis and meiosis. Polymerase works by binding to a single DNA strand and creating the complementary strand. In the first of many original processes, the enzyme was used in vitro (in a controlled environment outside an organism). The double-stranded DNA was separated into two single strands by heating it to 94°C (201°F). At this temperature, however, the DNA polymerase used at the time were destroyed, so the enzyme had to be replenished after the heating stage of each cycle. The original procedure was very inefficient, since it required a great deal of time, large amounts of DNA polymerase, and continual attention throughout the process.
Later, this original PCR process was greatly improved by the use of DNA polymerase taken from thermophilic bacteria grown in geysers at a temperature of over 110°C (230°F). The DNA polymerase taken from these organisms is stable at high temperatures and, when used in PCR, does not break down when the mixture was heated to separate the DNA strands. Since there was no longer a need to add new DNA polymerase for each cycle, the process of copying a given DNA strand could be simplified and automated.
One of the first thermostable DNA polymerases was obtained from Thermus aquaticus and was called "Taq." Taq polymerase is widely used in current PCR practice. A disadvantage of Taq is that it sometimes makes mistakes when copying DNA, leading to mutations (errors) in the DNA sequence, since it lacks 3'→5' proofreading exonuclease activity. Polymerases such as Pwo or Pfu, obtained from Archaea, have proofreading mechanisms (mechanisms that check for errors) and can significantly reduce the number of mutations that occur in the copied DNA sequence. However these enzymes polymerise DNA at a much slower rate than Taq. Combinations of both Taq and Pfu are available nowadays that provide both high processivity (fast polymerisation) and high fidelity (accurate duplication of DNA).
PCR has been performed on DNA larger than 10 kilobases, however the average PCR is only several hundred to a few thousand bases of DNA. The problem with long PCR is that there is a balance between accuracy and processivity of the enzyme. Usually, the longer the fragment, the greater the probability of errors.
Patent wars
The PCR technique was patented by Cetus Corporation, where Mullis worked when he invented the technique in 1983. The Taq polymerase enzyme is also covered by patents. There have been several high-profile lawsuits related to the technique, including an unsuccessful lawsuit brought by DuPont. The pharmaceutical company Hoffmann-La Roche purchased the rights to the patents in 1992 and currently holds those that are still protected.
A related patent battle over the Taq polymerase enzyme is still ongoing in several jurisdictions around the world between Roche and Promega. Interestingly, it seems possible that the legal arguments will extend beyond the life of the original PCR and Taq polymerase patents, which expire in 2006.
References
- ^ The history of PCR: Smithsonian Institution Archives, Institutional History Division. Retrieved 24 June 2006.
- Sambrook, Joseph (2001). Molecular Cloning: A Laboratory Manual (3rd ed. ed.). Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press. ISBN 0-87969-576-5.
{{cite book}}:|edition=has extra text (help); Unknown parameter|coauthors=ignored (|author=suggested) (help) - Mullis, Kary (1998). Dancing Naked in the Mind Field. New York: Pantheon Books. ISBN 0-679-44255-3.
- Rabinow, Paul (1996). Making PCR: A Story of Biotechnology. Chicago: University of Chicago Press. ISBN 0-226-70146-8.
External links
- PCR Polymerase Chain Reaction (Animation)
- PCR - Polymerase Chain Reaction Articles, news, bioinformatics, and protocols for PCR.
- PCR Interactive Animation
- Online simulation of PCR processes against sequenced prokaryotes.
- Shockwave Animation of PCR by Dolan DNA Learning Center.
- The PCR Jump Station Information and links on the polymerase chain reaction
- PCR Narrated flash animation
