Antisense RNA
The antisense RNA ( aRNA ), also natural antisense transcript (NAT) called, is a single-stranded RNA that is complementary protein coding for a messenger RNA ( mRNA ) is. The aRNA is assigned to the lncRNAs or ncRNAs. It plays an important role in regulatory processes in cells, but it is also increasingly used in research as a tool for gene knockdown .
structure
The structure of aRNA is naturally similar to that of mRNA. However, almost all aRNAs have secondary structures , such as stem-loops and in some cases also more complex tertiary structures , such as pseudo-knots between these very same secondary structures. These structural elements determine the rate of degradation by intracellular ribonucleases and also the rate at which the aRNA pairs with the complementary RNA.
Action path
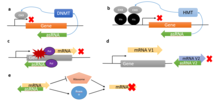
The aRNA can act in a number of ways. It mostly works by preventing the translation of a gene. But it can also have an epigenetic or activating effect.
Translation
The mRNA is transcribed from the template strand of the DNA . The non-template strand is thus the codogenic strand. If this - the strand complementary to the template strand - is also transcribed, an aRNA which is complementary to the mRNA is created. By base pairing with the complementary mRNA, the aRNA inhibits its translation in the cell .
An alternative mode of action is the binding of the aRNA to the binding sites of miRNA and the associated stabilization of the RNA. It is believed that this mechanism also plays a role in Alzheimer's disease . Thus the will expression of individual genes regulated .
Epigenetics
It is known that aRNA can also act epigenetically. Longer aRNAs can usually be observed here. This can be caused, for example, by the aRNA-induced methylation of CpG islands in the genome. Such an effect has been demonstrated , for example, in connection with the disease α-thalassemia or the silencing of X chromosomes .
activation
It is also possible that aRNA has an activating effect. This is achieved, for example, in that the aRNA binds to the RNA of a hairpin structure and thus exposes a Shine-Dalgarno sequence hidden in it and thus enables translation.
classification
Antisense RNA can represent both a cis and a trans-acting element. In the first case, the aRNA is transcribed from the complementary DNA strand. So one often has a very high degree or complete complementarity and the aRNA only has a single target RNA. In the latter case, the aRNA comes from a more distant gene. They usually show a lower degree of complementarity, which is why the complexes formed are more unstable and in some cases chaperones are required to stabilize the complex and several RNAs can be the target.
In addition to this classification, classification can also be made either according to the type of interaction (RNA-DNA, RNA-RNA or RNA-protein), the length of the aRNA (the limit is drawn here at 100 bp) or according to the type of promoter concerned. Finally, a classification is sometimes made according to the half-life of the aRNA in the cell, even if the importance of this is subordinate to that of the mating rate between aRNA and mRNA.
Use & occurrence
Antisense-RNA represents a natural possibility of gene regulation of protein biosynthesis . In humans there are at least 1600 antisense genes, for example the insulin-like growth factor 2 receptor (IGF-2). In the case of this gene, depending on its genetic makeup , a second promoter can be active at the 3 'end of the gene , via which antisense RNA is transcribed. This then prevents the translation of both alleles of this gene. The phenotype does not follow the Mendelian genetics . Antisense transcripts occur in more than 70% of the genes in cDNA databases (Fantom-3, GenBank).
Antisense RNA is used, for example, in biotechnology , for example in the commercially less successful Flavr-Savr tomato . Here, an artificial gene was introduced into the tomato that produces antisense RNA against a gene involved in the ripening process that codes for the enzyme polygalacturonase . This can delay the ripening process of the so-called Flavr-Savr tomato. Another example is the Amflora potato , where the technique was used to suppress amylose production in the potato.
In addition to its use in biotechnology, the antisense RNA system is increasingly being used in medicine and pharmacology. The first drug based on antisense technology and approved for sale is the antiviral drug fomivirsen against the cytomegalovirus . C. Frank Bennett (neurodegenerative diseases) and Adrian R. Krainer ( spinal muscular atrophy ) received the Breakthrough Prize in Life Sciences for the development of antisense RNA drugs in 2019 .
In such therapeutic approaches, attempts are usually made to shut down a gene using aRNA. There are two approaches to this. On the one hand, an aRNA for a type of anti-gene can be generated that then binds to the mRNA of the target gene. Alternatively, a 15 to 20 bp long aRNA can be used that targets a specific sequence, which is usually sufficient. One possibility here is to attack the 5 'end of the mRNA and thus completely prevent translation. It is just as easy to bind to any other point within the mRNA, which usually has the same effectiveness.
In molecular and cell biology , in vitro generated antisense RNA is used for in situ hybridizations .
literature
- ↑ Farzaneh Modarresi, Mohammad Ali Faghihi, Miguel A Lopez-Toledano, Roya Pedram Fatemi, Marco Magistri: Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation . In: Nature Biotechnology . tape 30 , no. 5 , May 2012, ISSN 1087-0156 , p. 453–459 , doi : 10.1038 / nbt.2158 , PMID 22446693 , PMC 4144683 (free full text).
- ↑ Jian-zhong Xu, Jun-lan Zhang, Wei-guo Zhang: Antisense RNA: the new favorite in genetic research . In: Journal of Zhejiang University SCIENCE B . tape 19 , no. October 10 , 2018, ISSN 1673-1581 , p. 739-749 , doi : 10.1631 / jzus.B1700594 , PMID 30269442 , PMC 6194357 (free full text).
- ↑ Yutaka Eguchi, Tateo Itoh, Jun-ichi Tomizawa: Antisense RNA . In: Annual Review of Biochemistry . tape 60 , no. 1 , June 1991, ISSN 0066-4154 , pp. 631-652 , doi : 10.1146 / annurev.bi.60.070191.003215 .
- ↑ João F. Menino, Agostinho J. Almeida, Fernando Rodrigues: Gene Knockdown in Paracoccidioides brasiliensis Using Antisense RNA . In: Host-Fungus Interactions . tape 845 . Humana Press, Totowa, NJ 2012, ISBN 978-1-61779-538-1 , pp. 187-198 , doi : 10.1007 / 978-1-61779-539-8_12 .
- ↑ a b R.W. Simons: Antisense RNA . In: Encyclopedia of Genetics . Elsevier, 2001, ISBN 0-12-227080-0 , pp. 83–84 , doi : 10.1006 / rwgn.2001.0061 ( elsevier.com [accessed October 21, 2019]).
- ^ A b E Wagner, S Altuvia, P Romby: 12 Antisense RNAs in bacteria and their genetic elements . In: Advances in Genetics . tape 46 . Elsevier, 2002, ISBN 0-12-017646-7 , pp. 361-398 , doi : 10.1016 / s0065-2660 (02) 46013-0 .
- ↑ Mohammad Ali Faghihi, Farzaneh Modarresi, Ahmad M. Khalil, Douglas E. Wood, Barbara G. Sahagan: Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of beta-secretase . In: Nature Medicine . tape 14 , no. July 7 , 2008, ISSN 1546-170X , p. 723-730 , doi : 10.1038 / nm1784 , PMID 18587408 , PMC 2826895 (free full text).
- ↑ Mohammad Ali Faghihi, Ming Zhang, Jia Huang, Farzaneh Modarresi, Marcel P. Van der Brug: Evidence for natural antisense transcript-mediated inhibition of microRNA function . In: Genome Biology . tape 11 , no. 5 , 2010, ISSN 1474-760X , p. R56 , doi : 10.1186 / gb-2010-11-5-r56 , PMID 20507594 , PMC 2898074 (free full text).
- ↑ Kevin V. Morris, Peter K. Vogt: Long antisense non-coding RNAs and their role in transcription and oncogenesis . In: Cell Cycle . tape 9 , no. July 13 , 2010, ISSN 1538-4101 , p. 2544-2547 , doi : 10.4161 / cc.9.13.12145 , PMID 20581457 , PMC 3040850 (free full text).
- ↑ Cristina Tufarelli, Jackie A Sloane Stanley, David Garrick, Jackie A Sharpe, Helena Ayyub: Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease . In: Nature Genetics . tape 34 , no. 2 , June 2003, ISSN 1061-4036 , p. 157-165 , doi : 10.1038 / ng1157 .
- ↑ Jeannie Lee, Lance S Davidow, David Warshawsky: Tsix, a gene antisense to Xist at the X-inactivation center . In: Nature Genetics . tape 21 , no. 4 , April 1999, ISSN 1061-4036 , pp. 400–404 , doi : 10.1038 / 7734 ( nature.com [accessed October 22, 2019]).
- ↑ Vicent Pelechano, Lars M. Steinmetz: Gene regulation by antisense transcription . In: Nature Reviews Genetics . tape 14 , no. December 12 , 2013, ISSN 1471-0056 , p. 880-893 , doi : 10.1038 / nrg3594 .
- ↑ Koji Numata, Yuki Okada, Rintaro Saito, Hidenori Kiyosawa, Akio Kanai: Comparative analysis of cis-encoded antisense RNAs in eukaryotes . In: Genes . tape 392 , no. 1-2 , May 2007, pp. 134–141 , doi : 10.1016 / j.gene.2006.12.005 ( elsevier.com [accessed October 20, 2019]).
- ↑ Fatemeh Saberi, Mehdi Kamali, Ali Najafi, Alavieh Yazdanparast, Mehrdad Moosazadeh Moghaddam: Natural antisense RNAs as mRNA regulatory elements in bacteria: a review on function and applications . In: Cellular & Molecular Biology Letters . tape 21 , no. 1 , December 2016, ISSN 1425-8153 , p. 6 , doi : 10.1186 / s11658-016-0007-z , PMID 28536609 , PMC 5415839 (free full text).
- ↑ Marco Magistri, Mohammad Ali Faghihi, Georges St Laurent, Claes Wahlestedt: Regulation of chromatin structure by long noncoding RNAs: focus on natural antisense transcripts . In: Trends in Genetics . tape 28 , no. 8 , August 2012, p. 389–396 , doi : 10.1016 / j.tig.2012.03.013 , PMID 22541732 , PMC 3768148 (free full text) - ( elsevier.com [accessed October 20, 2019]).
- ↑ SK Ballas, WC Sherwood: Rapid in vivo destruction of Yt (a +) erythrocytes in a recipient with anti-Yta . In: transfusion . tape 17 , no. January 1 , 1977, ISSN 0041-1132 , pp. 65-66 , doi : 10.1046 / j.1537-2995.1977.17177128889.x , PMID 841677 .
- ^ RIKEN Genome Exploration Research Group and Genome Science Group (Genome Network Project Core Group) and the FANTOM Consortium: Antisense Transcription in the Mammalian Transcriptome . In: Science . tape 309 , no. 5740 , September 2, 2005, ISSN 0036-8075 , p. 1564–1566 , doi : 10.1126 / science.1112009 .
- ^ RA Sanders, W. Hiatt: Tomato transgene structure and silencing. In: Nat. Biotechnol. Vol. 23, 2005, pp. 287-289. PMID 15765076
- ↑ BASF: Amflora ( page no longer available , search in web archives ) Info: The link was automatically marked as defective. Please check the link according to the instructions and then remove this notice.
- ↑ David P. Clark, Nanette J. Pazdernik: Inherited Defects and Gene Therapy . In: Biotechnology . Elsevier, 2016, ISBN 978-0-12-385015-7 , pp. 523-564 , doi : 10.1016 / b978-0-12-385015-7.00017-x .
See also
Web links
- Hamburger Bildungsserver: A genetically modified food: The Flavr Savr tomato
- www.transgen.de: Changed plant development: Mature - and yet more durable. ( Memento of December 10, 2008 in the Internet Archive ) of December 10, 2008.