Triosephosphate isomerase
| Triosephosphate isomerase | ||
|---|---|---|
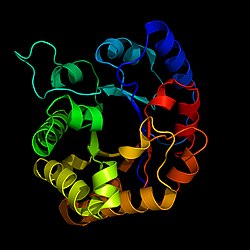
|
||
| Belt model according to PDB 2jk2 | ||
| Properties of human protein | ||
| Mass / length primary structure | 248 amino acids | |
| Secondary to quaternary structure | Homodimer | |
| Isoforms | 2 | |
| Identifier | ||
| Gene name | TPI1 | |
| External IDs | ||
| Enzyme classification | ||
| EC, category | 5.3.1.1 , isomerase | |
| Substrate | Dihydroxyacetone phosphate (= glyceron phosphate) | |
| Products | D-glyceraldehyde-3-phosphate | |
| Occurrence | ||
| Homology family | CLU_024251_2_0 | |
| Parent taxon | Chordates | |
| Orthologue | ||
| human | House mouse | |
| Entrez | 7167 | 21991 |
| Ensemble | ENSG00000111669 | ENSMUSG00000023456 |
| UniProt | P60174 | P17751 |
| Refseq (mRNA) | NM_000365 | NM_009415 |
| Refseq (protein) | NP_000356 | NP_033441 |
| Gene locus | Chr 12: 6.87 - 6.87 Mb | Chr 6: 124.81 - 124.81 Mb |
| PubMed search | 7167 |
21991
|
The triose phosphate isomerase ( TIM, TPI ) is the enzyme that dihydroxyacetone phosphate (DHAP) to glyceraldehyde-3-phosphate is converted (CAP). This is a sub-step of glycolysis . TPI is therefore indispensable for all living things that can only utilize glucose or fructose by means of glycolysis.
In humans, a gene (TPI1) on chromosome 12 , locus 12p13 codes for the functional protein, at least three pseudogenes are known. Mutations in the gene can cause triose phosphate isomerase deficiency .
A potent inhibitor is 2-phosphoglycolate , which is broken down in plants during photorespiration .
structure
The triosephosphate isomerase is a member of the all-α- and all-β-class (α / β) of proteins and a homodimer, which consists of two sequence-identical subunits (chains) each with 247 amino acids. Each TPI monomer (chain) contains the full set of catalytic amino acid residues, but the enzyme is only active in the oligomeric form. Therefore, dimerization is essential for the enzyme to function fully, although it is not believed that there is cooperativity between the two active sites.
Each subunit contains 8 outer α-helices that 8 inner β-strands surrounded and a conserved structural domain form, which as a closed α / β-barrel said (α / β) or more accurate than TIM barrel (engl. TIM barrel is) denotes . The TIM barrel was originally named after the enzyme and is estimated to be contained in 10% of all enzymes. Most TIM barrel domains are characterized by the presence of the active site of the enzyme in the regions of the lower loop created by the eight loops that connect the C -termini of the β-strands with the N -termini of the α-helices connect. TIM barrel proteins also share a structurally conserved phosphate binding motif with the phosphate group located in the substrate or in the cofactors.
In each chain, non-polar amino acids that point inwards from the β-strands contribute to the hydrophobic core of the structure. The α-helices are amphipathic : their outer surfaces (which come into contact with water) are polar, while their inner surfaces are largely hydrophobic. The loops are a mixture of polar and non-polar amino acid residues.
Catalyzed equilibrium
TPI creates a balance between the intermediates dihydroxyacetone phosphate (DHAP) and glyceraldehyde-3-phosphate (GAP). The substrates are formed from fructose-1,6-bisphosphate in the upstream aldolase reaction. The balance is strongly on the DHAP side, but GAP is required for the glycolysis to continue, so that the balance shifts when the product is withdrawn ( Le Chatelier's principle ).
The catalysis takes place via an enediol or enediolate intermediate. Here enters glutamate residue (Glu 165 in the active center of the enzyme with the unusually high pK) s value of 6.5 as a base on a histidine residue (His 95 ) as the acid .
The conversion of DHAP to GAP requires a fancy reaction mechanism , in the course of which Glu 165 abstracts an H + ion from C atom 2, while His 95 releases an H + ion to the C1 atom. Since the carboxy group of Glu is much more acidic than the C2 atom, this mechanism can not take place under non-enzymatic conditions. The TIM , on the other hand, forms what are known as low-barrier hydrogen bonds ( LBHB ), a special type of hydrogen bond with −40 to −80 KJ / mol (instead of around −12 up to −30 kJ / mol) are significantly more stable. These LBHB are achieved by simultaneous protonation and deprotonation at the carbon atoms C1 and C2.
A 10 amino acid long sequence of the enzyme, a so-called loop, covers the active center when the substrate is loaded. On the one hand, this stabilizes the enediol intermediate and increases the catalytic activity by a factor of 10 5 ; on the other hand, this intermediate product is prevented from escaping - the enediol phosphate would spontaneously dephosphorylate and rearrange itself to the toxic methylglyoxal .
TIM is considered to be the " catalytically perfect enzyme ". This means that changes to the enzyme, regardless of the type, are no longer able to bring about an increase in turnover: The turnover rate of 4300 substrate molecule turnover per second is only limited by the diffusion speeds of substrate and product.
Web links
Individual evidence
- ↑ Anderson, LE. (1971): Chloroplast and cytoplasmic enzymes. II. Pea leaf triose phosphate isomerases . In: Biochim Biophys Acta . 235 (1); 237-244; PMID 5089710 ; doi : 10.1016 / 0005-2744 (71) 90051-9 .
- ↑ C. Rodríguez-Almazán, R. Arreola, D. Rodríguez-Larrea, B. Aguirre-López, MT de Gómez-Puyou, R. Pérez-Montfort, M. Costas, A. Gómez-Puyou, A. Torres-Larios : Structural basis of human triosephosphate isomerase deficiency: mutation E104D is related to alterations of a conserved water network at the dimer interface. In: Journal of Biological Chemistry . Volume 283, Number 34, August 2008, pp. 23254-23263, doi : 10.1074 / jbc.M802145200 , PMID 18562316 .
- ↑ KD Schnackerz, RW Gracy: Probing the catalytic sites of triosephosphate isomerase by 31P-NMR with reversibly and irreversibly binding substrate analogues. In: European Journal of Biochemistry . Volume 199, Number 1, July 1991, pp. 231-238, doi : 10.1111 / j.1432-1033.1991.tb16114.x , PMID 2065677 .
- ↑ A. Marchler-Bauer, Y. Bo, L. Han, J. He, CJ Lanczycki, S. Lu, F. Chitsaz, MK Derbyshire, RC Geer, NR Gonzales, M. Gwadz, DI Hurwitz, F. Lu, GH Marchler, JS Song, N. Thanki, Z. Wang, RA Yamashita, D. Zhang, C. Zheng, LY Geer, SH Bryant: CDD / SPARCLE: functional classification of proteins via subfamily domain architectures. In: Nucleic acids research. Volume 45, D101 2017, pp. D200 – D203, doi : 10.1093 / nar / gkw1129 , PMID 27899674 , PMC 5210587 (free full text).
- ↑ E. Lolis, GA Petsko: Crystallographic analysis of the complex between triosephosphate isomerase and 2-phosphoglycolate at 2.5-A resolution: implications for catalysis. In: Biochemistry. Volume 29, Number 28, July 1990, pp. 6619-6625, doi : 10.1021 / bi00480a010 , PMID 2204418 .


