trp operon
The tryptophan operon , short trp operon , is an operon that is used for the intracellular synthesis of tryptophan in bacteria such as. B. Escherichia coli plays a central role.
This operon is active when the cell has to produce tryptophan itself because this amino acid is not sufficiently present in the surrounding medium. In contrast, if the intracellular concentration of tryptophan is sufficiently high, the operon is usually inactivated by a specific repressor .
The structural genes of the operon code for polypeptides of enzymes with the help of which the amino acid tryptophan can be produced from a precursor ( chorismic acid ) in several steps. The access to these genes, which is necessary for the protein biosynthesis of the enzymatically active proteins, is regulated via upstream regulatory regions in the operon by preventing or interrupting transcription here .
trp operon in E. coli
Structure of the operon
The trp operon in Escherichia coli bacteria, like their lac operons and other operons in the genome of prokaryotic cells, consists of functionally different DNA segments. In addition to the structural genes as such DNA segments whose base sequence determines the structure of enzymatically active proteins when lives (RNA transcribed ) and subsequently translated (in amino acid sequence translated ), the operon contains other DNA segments as regulatory regions which are important for the regulation of transcription.
Next to the promoter region (P), to which an RNA polymerase molecule can bind for transcription, is the operator region (O), to which a repressor protein can bind and thus prevent the start of transcription ( repression ). In the untranslated area that follows, there is another regulatory region in the leader sequence ( trpL ), with which a transcription process that has started can be delayed or terminated ( attenuation ). Only then do the sections of the structural genes ( trpE, trpD, trpC, trpB, trpA ) with open reading frames , which encode various polypeptides for several proteins , follow downstream . When these are formed together, they can catalyze a sequence of reaction steps in tryptophan biosynthesis . The polycistronic mRNA required for this is provided by the trp operon - provided that the transcription can be started and not terminated.
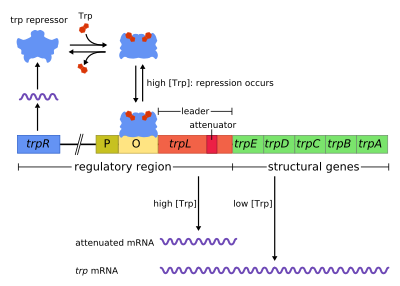
(P, O, trpL are as regulatory DNA region before the structural genes trpE, trpD, trpC, trpB, trpA downstream)
related to additional regulatory elements
( trpR gene for trp repressor protein, which with tryptophan (Trp) activated as a corepressor can bind to the operator O and thus prevents the start of transcription at the promoter P).
Modes of regulation
The transcription of the genes by RNA polymerase is a function of the tryptophan concentration regulated , primarily via the operator by repression . A blocking protein can bind to the operator area as a repressor - but only if it has previously bound a molecule of the amino acid tryptophan as a ligand . If there is no tryptophan that could form this reversible bond, the repressor remains inactive and the operon is not repressed. If, on the other hand, there is enough tryptophan so that it is bound as a cofactor to the protein or as a corepressor to the aporepressor , the repressor protein is activated and can thus bind to the operator, which then blocks transcription.
An mRNA is required for the biosynthesis of this repressor , which is encoded by the associated regulatory gene trpR at another point on the DNA strand.
With the present repressor, the cell's own tryptophan formation can now be regulated automatically depending on the tryptophan concentration in the cytosol, according to the principle of negative feedback . Since tryptophan is also the product of the regulated manufacturing process here, one speaks of an end product repression: the product of a synthesis itself has an indirect effect on this by hindering further gene expression for the enzymes involved, repressing it.
In addition to this primary mode of regulation of repression via the operator area, with which a start of transcription can be suppressed, a second additional mode of regulation of attenuation is known for the trp operon , with which a transcription that has started can be prematurely terminated. Further regulatory sequences in the untranslated area ( 5'-UTR ) play a role here; in addition to - a so-called leader peptide coding - trpL sequence there are those different alternative folds in the resulting mRNA permit. One of these hairpin structures represents the so-called termination signal of the attenuator sequence: if it is formed, the RNA strand that has formed up to that point is separated from the RNA polymerase and the latter from the DNA, which terminates the transcription. This does not happen if the concentration of tRNA (tRNA Trp ) loaded with tryptophan is very low - i.e. only when the tryptophan level is high. In this way, a finer adjustment of the gene expression to the cytosolic tryptophan concentration is possible, and an excessive tryptophan synthesis is also counteracted.
Coded enzymes
The structural genes for tryptophan biosynthesis in the trp operon of the bacterium Escherichia coli are located downstream of the leader sequence trpL and comprise seven segments of trpEGDCFBA in five sections,
- trpE
- trpD (with G)
- trpC (with F)
- trpB
- trpA
each of the seven genetic segments coding for an amino acid sequence in the polypeptide which, as a protein domain , catalyzes one of the reaction steps in the synthesis of tryptophan from chorismate . Chorismate is provided via the shikimic acid route .
Under control of the trp operon are its structural genes by RNA polymerase together as a polycistronic mRNA transcribed , followed by ribosomes into polypeptides translated . Since the sequences of trpG with trpD as well as those of trpF with trpC are each in the same open reading frame, these two sections each code only for a bifunctional polypeptide which contains two different enzymatically active domains.
In contrast, between the five separately listed sections there are four short intercistronic areas. In addition to a stop codon and a start codon as the end of the previous and the beginning of the next reading frame, the ribosomal binding site (RBS) with the Shine-Dalgarno sequence that a ribosome needs to attach to the mRNA strand can be found here. Only then can the coding nucleotide sequence of a reading frame be translated into the amino acid sequence of a polypeptide.
After the translational folding a naszierndes polypeptide in the aqueous salt-cell environment to a native protein of certain shape , and can associate with other molecules. In this way, the proteins of the polypeptide types trp-E and trp-D (with G) often attach to one another and form protein complexes consisting of four subunits, just as those of trp-B and trp-A two plus two form a heterotetramer . These multi-enzyme complexes unfold their subunits for different chemical reactions of the biosynthetic pathway as catalytic activity
- Anthranilate synthase (trp-G and trp-E): chorismate → → anthranilate
- Anthranilate phosphoribosyl transferase (trp-D): Anthranilate → N- (5-phosphoribosyl) anthranilate
- Phosphoribosyl anthranilate isomerase (trp-F): N- (5-phosphoribosyl) anthranilate → 1- (o-carboxyphenylamino) -1-deoxyribulose-5-phosphate
- Indole glycerol phosphate synthase (trp-C): 1- (o-carboxyphenylamino) -1-deoxyribulose-5-phosphate → indole-3-glycerol phosphate
- Tryptophan synthase (trp-A and trp-B): indole-3-glycerol phosphate ↔ indole → L- tryptophan
With their help, L- Triptophan can be built up from chorismic acid . Only one of these steps is reversible (↔).
Individual evidence
- ↑ C. Yanofsky, T. Platt, IP Crawford, BP Nichols, GE Christie, H. Horowitz, M. VanCleemput, AM Wu: The complete nucleotide sequence of the tryptophan operon of Escherichia coli. In: Nucleic acids research. Volume 9, Number 24, December 1981, pp. 6647-6668, PMID 7038627 , PMC 327632 (free full text) (review).
- ↑ a b c C. Yanofsky: RNA-based regulation of genes of tryptophan synthesis and degradation, in bacteria. In: RNA. Volume 13, Number 8, August 2007, pp. 1141–1154, Figure 2. doi: 10.1261 / rna.620507 , PMID 17601995 , PMC 1924887 (free full text) (review).
- ↑ see entry UniProtKB - P0AD92 (LPW_ECOLI) in the bioinformatic database for proteins UniProt .
literature
- Nancy Trun, Janine Trempy: Gene expression and regulation. In: Nancy Trun, Janine Trempy: Fundamental Bacterial Genetics. Blackwell, Malden MA et al. a. 2004, ISBN 0-632-04448-9 , pp. 191-212, online (PDF; 500 kB) .
