Attenuation (gene expression)
Transcriptional attenuation is a form of regulation of gene expression in prokaryotes with which a transcription that has started can be prematurely terminated.
The transcription process is terminated in that the transcribing RNA polymerase is separated from the DNA template if, as a result of interactions within the mRNA starting area that has just been built up, a hairpin structure is formed by internal base pairing at the terminator site, the attenuator . However, this terminating loop formation in the secondary structure of the mRNA is not possible in certain positions of a ribosome that is advancing simultaneously and translating the resulting mRNA . The ribosome only assumes these positions in the event of a delayed translation; it is only when this is delayed that the transcription can continue beyond the attenuator area. The translation process is delayed, for example, if a tRNA loaded with the specific amino acid is not promptly available because the amino acid in question is only present in the cell in low concentrations. Under such conditions a complete transcription can take place - for example of genes for the synthesis of an amino acid, if there is a lack of this.
The prerequisite for this regulatory mechanism is that the processes of transcription and translation take place together and almost synchronously, i.e. that the RNA polymerase is still transcribing and building the mRNA molecule, while a ribosome is already sitting at another point of the same mRNA molecule and translating at the same time. This temporal and spatial coupling is only given in prokaryotes. In eukaryotic cells, the transcription takes place in the cell compartment of the cell nucleus , then the mRNA is exported from the caryoplasm and the translation takes place outside the nucleus in the cytoplasm , separated in time and space.
Example: Tryptophan operon in Escherichia coli
The trp operon in bacteria such as Escherichia coli is a classic example of transcriptional attenuation as an additional mode of gene regulation, first described by Charles Yanofsky in 1981 . But access to the operon is primarily regulated by repression in E. coli as well . A trpR gene codes for a repressor protein that reversibly binds to an operator ( O ) located in the promoter region ( P ) and thus obstructs the access for the RNA polymerase if it was previously activated as a corepressor by the amino acid tryptophan (Trp) . In this way, the transcription is regulated with negative feedback depending on the intracellular tryptophan level and is suppressed as the level rises. With a high tryptophan concentration , the trp operon is therefore repressed with a very high degree of probability and transcription hardly takes place.
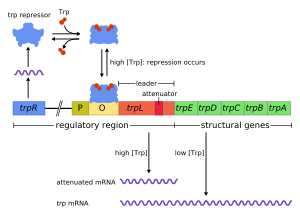
However, if transcription does get under way, the attenuation represents an additional safeguarding mechanism with which a premature termination can be forced at high levels of tRNA (tRNA Trp ) loaded with tryptophan - even before the subsequent structural genes of the operon are transcribed. These successive genes ( trpE, trpG-D, trpC-F, trpB and trpA ) code for seven polypeptides that are needed in enzymatically active proteins for the biosynthesis of tryptophan - if there is a deficiency in this amino acid. In front of the open reading frames of these genes there is now a so-called untranslated area , the 5'-UTR or leader sequence , with those sequences that make an attenuation possible during the formation of the mRNA.
In this area of around 160 nucleotides at the 5 'start of the mRNA there are four shorter palindromic sequences : each about a dozen nucleotides long single-stranded sequences (1st, 2nd, 3rd, 4th) that can complement each other and in this way, by pairing bases with one another, they are able to form double-stranded sections of a hairpin structure in the mRNA. Different stem-loop formations are possible (1: 2, 2: 3, 3: 4) with complementary palindromes . A formation of the rear (3: 4) mRNA loop leads - because of the strong bonds close to the loop via hydrogen bonds (GCGGGC ... in 4.) - to the stop of the RNA polymerase molecule, and - because of the subsequent weak bonds (.. .UUUUUU) - to detach from the DNA strand, with which the transcription is terminated prematurely, even before the first gene of the operon ( trpE ) has been reached. The structure-bearing sequence required for this Rho factor-independent termination is referred to as the (intrinsic) termination signal of the attenuator sequence .
The preceding sequences allow an alternative loop formation within the resulting mRNA transcript, with which the premature termination of transcription can be avoided. Because if the sequence 2 is paired with 3 (2: 3), the pairing of 3 with 4 is not possible. The formation of this loop structure 2: 3 only occurs when the ribosome stays in sequence 1, which is initially synchronized and follows the RNA polymerase, when it is clearly delayed.
This decisive delay can occur because sequence 1 also represents the end of another regulatory section, the trpL sequence, a section with 14 codons - which is rather unusual in a 5 'UT region . The last five of these before the stop codon , including twice the codon for tryptophan , are in sequence 1. The ribosome now translates this coding nucleotide sequence trpL of the mRNA into a so-called leader peptide , and then stays here a little longer when loaded tRNA Trp for the Trp tandem is not available promptly, for example in the case of tryptophan deficiency in the cytosol. At the same time, however, sequence 1 as a possible partner for sequence 2 is withdrawn, which then couples with 3 (2: 3), which means that 3: 4 is no longer possible. The premature termination of the transcription is thus undermined.
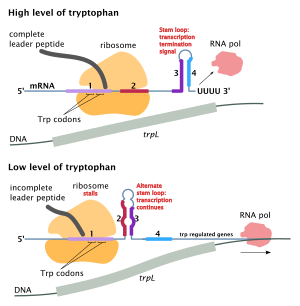
If the Trp level is low , the formation of the leader peptide in region 1 is delayed, which makes the structure 2: 3 possible and thus the complete transcription .
If, on the other hand, there is enough tryptophan, a ribosome can quickly translate this area of the leader sequence and quickly follow the polymerase via sequence 2. No trunk-loop structure 2: 3 is then formed; the sequence 3 can therefore form the structure 3: 4 with 4 as soon as it is present: the termination signal for the RNA polymerase interrupts the transcription (and the genes trpE to trpA are not transcribed).
If too little tryptophan is available, the ribosome needs significantly more time to translate sequence 1 into the leader peptide. The delayed translation leads to the stem-loop structure 2: 3 in the transcript created downstream, thus allowing the complete transcription of all genes trpE to trpA by the RNA polymerase into a polycistronic mRNA of around 6800 nucleotides - and thus enables the subsequent translation of this mRNA through ribosomes into the gene products TrpE to TrpA: as enzymatically active proteins, they enable increased tryptophan synthesis, which is advantageous in the case of tryptophan deficiency.
Whether the attenuation occurs or the strand is not withdrawn from the polymerase is decided on the migration of the first ribosome through the space of the folding possibilities at the beginning of the nascent mRNA. The unusual task of building up a short tryptophan-containing peptide in the so-called untranslated area becomes a criterion for the course of transcription and makes the expression of the trp genes dependent on the current Trp concentration in the cell environment. Only when the level is low is there a brief pause that makes successful transcription possible.
Further examples of transcriptional attenuation
A number of other operons are now known that use transcriptional attenuation as a mode of regulation. In addition to operons for amino acid biosynthesis, for example of phenylalanine , histidine , leucine , threonine , methionine or cysteine, there are also operons for amino acid degradation, and some with modifications of the attenuation mechanism described above.
For example, the tna operon in E. coli contains the genes that code for enzymes that break down tryptophan (tryptophanase) and are mainly transcribed in the cell when there is an excess of tryptophan. Under these conditions, a translated tryptophan-containing leader peptide prevents the premature termination, which is dependent on a Rho factor, by blocking the binding sites in the leader region that are necessary for this.
The expression of the operon pyrBI in E. coli , which codes for an enzyme involved in the biosynthesis of pyrimidine , is primarily regulated by attenuation. Here it is the polymerase that pauses when a pyrimidine triphosphate ( UTP ) is not readily available in a certain region of the leader sequence, thereby influencing the position and movement of the ribosome in such a way that it prevents the formation of the terminator signal. In the pyr operon in Bacillus subtilis , it is a special RNA-binding protein, PyrR, which is activated by a pyrimidine monophosphate ( UMP ) and can then bind to the mRNA in such a way that the intrinsic terminator is created; on the other hand, if the UMP level is low, it remains inactive, so that an alternative mRNA structure can be folded that enables complete transcription.
In the trp operon in B. subtilis , the expression of its six structural genes trpEDCFBA is regulated by a typophane-activated RNA-binding protein ( TRAP ), which binds to the alternative structural sequence in the leader region and indirectly induces the formation of the terminator signal and thus becomes transcriptional Attenuation leads. In addition, the mRNA can be refolded over a hundred nucleotides further downstream, with which the ribosome binding site is embedded in a hairpin structure in front of trpE , so that the initiation of translation of this gene is blocked. TRAP is a toroidal complex of 11 identical protein subunits, each of which forms a niche for tryptophan binding between them. If these are occupied, the mRNA sequence, which contains 11 (U / G) AG triplets in series with 2/3 nucleotides apart, can be placed peripherally around the outer surface of the structure around a tire and is made up of 11 KKR motifs ( Lys -Lys- Arg ) recognized opposite.
Transcriptional attenuation is therefore a not uncommon form of regulation of gene expression in prokaryotic cells, in which alternative folding patterns in the newly formed initial region of an mRNA offer the possibility of prematurely terminating a transcription that has started, depending on the currently prevailing metabolic conditions in the cell - even before structural genes transcribed into mRNA - or not.
Characteristics of regulation via attenuation
In addition to repression, the possibility of attenuation represents an additional mode of regulation of gene expression. In the meantime, various modifications are known with which it can be implemented.
In the case of the trp operon in E. coli , the limits of the control range for the structural genes expressed depending on the cellular tryptophan concentration with the repressor present are about a factor of 700 apart. Even when the repressor gene trpR is switched off , a tryptophane-regulated attenuation of gene expression by a factor of about 10 is still possible through attenuation alone.
Requirements for this attenuation are
- simultaneous transcription and translation with
- synchronized procession of RNA polymerase and ribosome
- with different folding possibilities of the nascent mRNA
- within the region of the 5 'UTR upstream of the structural genes
- with interaction between the newly created region and previously created,
- with which a structure formation is intrinsically possible, which acts as a termination signal,
- to which, however, alternative structure formations are possible, which exclude this,
- where there are also further regulatory regions or regulatory elements,
- by means of which the choice between the alternatives is decisively influenced,
- and this now depending on the currently prevailing metabolic conditions,
- where the variability of such a variable then primarily plays a role,
- which is linked to the gene product or its effect.
Remarks
- ↑ The process of synchronizing the transcribing RNA polymerase with the translating ribosome is achieved by pausing the polymerase 1: 2 after the signaling mRNA structure has formed - and waiting until the first ribosome begins to translate, whereby this hairpin structure is then dissolved.
- ↑ One seventh of the amino acids of this peptide with the amino acid sequence MKAIFVLKGWWRTS stop (see LPW ECOLI in UniProt database) is therefore tryptophan (W). But this amino acid is usually a very seldom used building block in proteins in E. coli, only around every hundredth.
Individual evidence
- ↑ Wilfried Janning, Elisabeth Knust: Genetics: General Genetics - Molecular Genetics - Developmental Genetics . 2nd Edition. Georg Thieme, Stuttgart 2008, ISBN 978-3-13-151422-6 , p. 211 ff .
- ^ Charles Yanofsky: Attenuation in the control of expression of bacterial operons . In: Nature . 289, No. 5800, February 1981, pp. 751-758. PMID 7007895 .
- ↑ a b c C. Yanofsky, T. Platt, I. Crawford, B. Nichols, G. Christie, H. Horowitz, M. Vancleemput, A. Wu: The complete nucleotide sequence of the tryptophan operon of Escherichia coli . In: Nucleic Acids Res . 9, No. 24, December 1981, pp. 6647-6668. PMC 327632 (free full text).
- ↑ a b c C. Yanofsky: RNA-based regulation of genes of tryptophan synthesis and degradation, in bacteria . In: RNA . 13, No. 8, August 2007, pp. 1141-1154. PMC 1924887 (free full text).
- ^ A b C. Yanofsky: Transcription Attenuation: Once Viewed as a Novel Regulatory Strategy . In: Journal of Bacteriology . 182, No. 1, January 2000, pp. 1-8. PMC 94232 (free full text).
- ↑ K. Konan, C. Yanofsky: Role of ribosome release in regulation of tna operon expression in Escherichia coli . In: Journal of Bacteriology . 181, No. 5, August 1999, pp. 1530-1536. PMC 93543 (free full text).
- ↑ J. Donahue, C. Turnbough: Nucleotide-specific transcriptional pausing in the pyrBI leader region of Escherichia coli K-12 . In: Journal of Biological Chemistry . 269, No. 27, July 1994, pp. 18185-18191. PMID 7517939 .
- ^ Y. Lu, R. Turner, R. Switzer: Function of RNA secondary structures in transcriptional attenuation of the Bacillus subtilis pyr operon . In: Proc Natl Acad Sci . 93, No. 25, October 1996, pp. 14462-14467. PMC 26155 (free full text).
literature
- Nancy Trun, Janine Trempy: Gene expression and regulation. In: Nancy Trun, Janine Trempy: Fundamental Bacterial Genetics. Blackwell, Malden MA et al. 2004, ISBN 0-632-04448-9 , online (PDF; 500 kB) .