5'-cap structure
The 5'-cap structure (from English cap 'cap') is a cap-like attachment at the 5 'end of mRNA molecules that is attached to the nucleus of eukaryotic cells . The 5 'cap not only protects an RNA from degradation, but is also important for the export of an mRNA from the nucleus into the cytoplasm and for the translation of the mRNA by ribosomes .
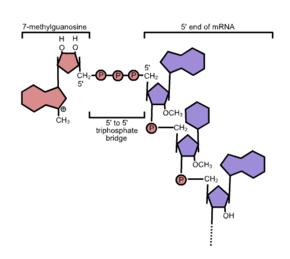
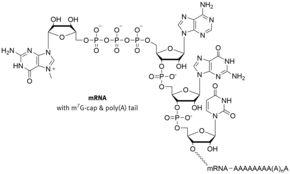
Often these are in the 5'-cap structure is a modified guanine - nucleotide that has a rare 5'-5 ' phosphodiester bond is linked to the head end of the RNA. This chemical reaction known as capping takes place during the transcription of a gene as soon as an RNA polymerase has linked the first nucleotides of an mRNA . In contrast to (kotranskriptionellen) capping the 5 'end which is polyadenylation of the 3'-end of - a mRNA tailing (from English tail , tail') said addition of a poly (A) tail - a post-transcriptional modification , the only after Separation of mRNA and RNA polymerase is performed.
In addition to mRNA, capping is also found in many non-coding RNAs . As an alternative to the cap structure, some RNA viruses use IRES or cap-independent translation elements to initiate translation .
Cap structures
Polymerase II
The classic m 7 G cap is found in mRNAs that are transcribed by RNA polymerase II . Here (after hydrolysis of the terminal γ-phosphate by the triphosphatase) a GMP residue (from GTP ) in the form of a 5'-5'-triphosphate bond is transferred to the 5'-end of the RNA by the so-called capping enzyme and then at position 7 of the guanine with the help of mRNA-cap-methyltransferase with consumption of S- adenosylmethionine (SAM), a universal methyl group donor, methylated , the result: 5'-5 'm 7 GpppN. In addition, further methylations of the first bases of the mRNA in position 2 ′ occur, one then speaks of Cap1 (only the first base methylated) or Cap2 (the first two bases methylated).
You can also find z. B. in trypanosomes far more complicated cap structures in which not only the first nucleotide is modified, but also the following ones (e.g. Trypanosoma brucei Cap-4).
Polymerase III
RNA polymerase III transcripts do not contain such 5′-cap structures, but a few RNAs have monomethylation on a γ-phosphate residue (e.g. U6 snRNA).
Viruses
Some viruses show a peculiarity in the biosynthetic pathway of the cap structure: GTP is first methylated and only then transferred to the RNA.
function
Like the poly (A) tail at the 3 ′ end of mRNAs, the cap structure plays an important role in stabilizing the mRNAs. Without this structure, the mRNAs in the cytoplasm are rapidly broken down by exonucleases from 5 ′ to 3 ′ .
The cap structure also plays an important role in the discharge of RNA from the cell nucleus through the nuclear pores into the cytoplasm (RNA export). It is bound by the cap-binding complex during transcription , which, in conjunction with other factors, ensures effective transport.
The cap structure is also of crucial importance for the initiation of translation . Both bound by CBC (during the first round of translation) and by eIF4E (during all further rounds) it ensures that the ribosome is recruited and begins with the initialization. This leads to a ring closure of the RNA (closed loop model of translation), in which the 5 ′ end interacts with the poly A tail (via eIF4E, eIF4G and the cytoplasmic poly A binding protein PABPC).
Since some viruses replicate exclusively in the cytoplasm, they do not receive a cap structure from the cellular machinery. To compensate for the disadvantages that this entails, they steal a cap from cellular mRNAs, known as cap snatching . An mRNA of the host organism is cleaved near the 5 'end (which of course has the cap structure) and used as a so-called capped leader to initiate viral translation.
The fact that viral polymerases do not produce m 7 G-Cap enables a specific differentiation between "foreign" and "endogenous" RNA - an RNA that only carries an unge cap tes triphosphate at the 5 'end can be used as an indication of a Infection apply. In fact, there is an intracellular receptor in the innate immune system (whose fundamental task is to differentiate between "foreign" and "self") with RIG-I , which recognizes precisely this structure as PAMP and subsequently triggers an antiviral immune response.
literature
- Rolf Knippers: Molecular Genetics. 8th revised edition. Georg Thieme Verlag, Stuttgart et al. 2001, ISBN 3-13-477008-3
- reactome: mRNA capping - doi : 10.3180 / REACT_1470.1
Individual evidence
- ↑ Hornung et al .: 5′-Triphosphate RNA Is the Ligand for RIG-I , In: Science, November 10, 2006: Vol. 314 no.5801 pp. 994-997 doi : 10.1126 / science.1132505 . Original publication in Science on the recognition of unge cap ter RNA