Lysine decarboxylase
| Lysine decarboxylase | ||
|---|---|---|

|
||
| Ribbon model of the constitutive lysine decarboxylase from E. coli , according to PDB 5FKZ | ||
| Identifier | ||
| External IDs |
|
|
| Enzyme classification | ||
| EC, category | 4.1.1.18 , lyase | |
| Response type | Decarboxylation | |
| Substrate | L- lysine | |
| Products | 1,5-diaminopentane + CO 2 | |
| Occurrence | ||
| Parent taxon | Bacteria , eukaryotes | |
Lysine decarboxylase ( LDC ) is an enzyme found mainly in bacteria and also in eukaryotes . LDC catalyzes the first step of quinolizidine alkaloid - biosynthesis , wherein the alkaloids with quinolizidine be synthesized -Grundstruktur, in particular lupine alkaloids .
Quinolizidine alkaloid biosynthesis
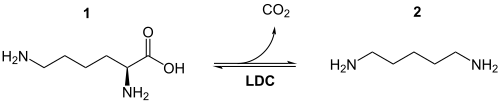
The first step in quinolizidine alkaloid biosynthesis consists of the splitting off of carbon dioxide ( decarboxylation ) from L- lysine ( 1 ) by means of lysine decarboxylase to cadaverine ( 2 ), also known as 1,5-diaminopentane.
Then cadaverine is converted to 3-aminopropanal with the help of the enzyme diamine oxidase . The synthesis of various quinolizidine alkaloids can only take place through the spontaneous cyclization of 3-aminopropanal to 1-piperidein . The alkaloids are then modified by dehydration , oxygenation or esterification . The formation of quinolizidine alkaloid esters marks the end of biosynthesis and the end products also represent a storage form for various organisms.
Among the Chinolizidinalkaloiden one distinguishes between Lupine alkaloids from the amino acid L -lysine apparent and more particularly in Lupine kinds (z. B. Lupinus angustifolius ), but also in the broom , Gorse and Goldregen occur and Nupharalkaloiden (from Nuphar ) formed on the Terpenweg are formed.
Well-known representatives of the lupine alkaloids are for example (-) - lupinine , (+) - epilupinine , (+) - multiflorine , (+) - lupanine , (+) - matrine , (-) - sparteine and (+) - cytisine .
Detection of the bacterial enzyme
Numerous types of bacteria have lysine decarboxylase. The detection of the enzyme in representatives of the gram-negative enterobacteria is used for differentiation and is part of a colored series to determine the genus or species . The test procedure was introduced in 1955 and has been part of miniaturized test systems since the 1970s (e.g. in the API 20 E system).
For the detection of the bacterial enzyme - often abbreviated as LDC - the standardized, lysine- containing nutrient medium is inoculated with bacterial material and incubated under anoxic conditions . To prevent oxygen from entering the test tube, the inoculated approach is covered with either paraffin oil or mineral oil , this prevents false-positive results. The formation of the diamine cadaverine increases the pH value in the test medium; the evaluation is based on the color change of the pH indicator integrated in the nutrient medium . For optimal enzyme activity a pH value below 5.5 is required (acidic range), while nutrient media usually have a neutral pH value.
Various test media
There are differences in the composition of the differentiation medium and the pH indicator used:
In the first developed nutrient medium according to Møller , D - glucose is used in addition to lysine , as well as bromocresol purple as a pH indicator, the pH value is adjusted to 6.0. The bacteria first utilize the small amount of glucose in a fermentation process , whereby acids are formed (compare mixed acid fermentation ), which lowers the pH value below 5.5. The reaction of the LDC that then sets in alkalizes the test medium and bromocresol purple indicates this by changing the color from yellow to purple . A comparison tube that does not contain lysine should always be carried so that the initial acid formation can be checked. A disadvantage is that the batch has to be incubated for four days before the evaluation can take place. A modification of this differentiation medium is the lysine iron agar , with which the formation of hydrogen sulfide by the bacteria can also be checked.
A method similar to that used for the detection of ornithine decarboxylase is considered a faster variant . The test medium does not contain any glucose; in addition to lysine, peptones and yeast extract are used, as well as bromothymol blue as a pH indicator, the pH value is adjusted to 5.2-5.4. The liquid nutrient medium is inoculated with copious amounts of bacterial material and incubated for four hours. If the bacteria have lysine decarboxylase, the test medium is made alkaline by the reaction product and bromothymol blue indicates this by changing the color from yellow to blue. A green color (pH value just below 7.0) is also rated as positive.
The substrate of the LDC test in the API 20 E system does not contain any glucose either, phenol red serves as a pH indicator and the original pH value is set to 6.2. An incubation period of 18 to 24 hours should be observed before the lysine decarboxylase reaction is assessed. The alkalization leads to a color change from yellow to red, an orange color (pH value just above 7.0) is also to be assessed as positive. If this is observed, an agreement of 98% with the Møller method results.
Examples of LDC positive and LDC negative bacteria
The detection of the bacterial lysine decarboxylase is important for the differentiation of the enterobacteria. Representatives of the genera Edwardsiella , Hafnia and Salmonella (except Salmonella enterica subsp. Enterica ser. Paratyphi) have this enzyme. In contrast , representatives of the genera Cedecea , Citrobacter , Moellerella , Proteus , Providencia , Rhanella , Shigella , Yersinia (except Yersinia ruckeri ) and the species Cronobacter sakazakii are LDC-negative. Within the genera Enterobacter , Escherichia , Klebsiella , Morganella and Serratia there are LDC-positive and -negative representatives, which the detection of the lysine decarboxylase reaction helps to distinguish.
Individual evidence
- ↑ UniProtKB results. In: UniProtKB. Accessed December 31, 2019 .
- ↑ a b Somnuk Bunsupa, Mami Yamazaki, Kazuki Saito: Quinolizidine alkaloid biosynthesis: recent advances and future prospects. In: Frontiers in Plant Science. Volume 3, 2012, doi: 10.3389 / fpls.2012.00239 .
- ↑ Quinolizidine alkaloid. In: Biocyclopedia. Accessed January 2, 2020 .
- ^ Lupine alkaloids. In: Lexicon of Biology. Spectrum, accessed January 2, 2020 .
- ↑ quinolizidine alkaloids. In: Lexicon of Biochemistry. Spectrum, accessed January 2, 2020 .
- ^ Ian Bass Seiple: The Lupine Alkaloids. (PDF) In: https://www.scripps.edu/ . Accessed January 2, 2020 .
- ↑ a b c Roland Süßmuth, Jürgen Eberspächer, Rainer Haag, Wolfgang Springer: Biochemical-microbiological internship . 1st edition. Thieme Verlag, Stuttgart / New York 1987, ISBN 3-13-685901-4 , p. 78-85 .
- ^ Vagn Møller: Simplified tests for some amino acid decarboxylases and for the arginine dihydrolase system . In: Acta pathologica et microbiologica Scandinavica . tape 36 , no. 2 , 1955, pp. 158-172 , doi : 10.1111 / j.1699-0463.1955.tb04583.x , PMID 14375937 .
- ↑ a b PB Smith, KM Tomfohrde, DL Rhoden, A. Balows: API system: a multitube micromethod for identification of Enterobacteriaceae . In: Applied Microbiology . tape 24 , no. 3 , September 1972, p. 449-452 , PMID 4562482 , PMC 376540 (free full text).
- ↑ a b c DC Brooker, ME Lund, DJ Blazevic: Rapid test for lysine decarboxylase activity in Enterobacteriaceae . In: Applied Microbiology . tape 26 , no. 4 , October 1973, p. 622-623 , PMID 4751806 , PMC 379861 (free full text).
- ^ A b c Elmer W. Koneman: Koneman's Color Atlas and Textbook of Diagnostic Microbiology . Lippincott Williams & Wilkins, 2006, ISBN 0-7817-3014-7 , pp. 225–226 ( limited preview in Google Book search).
- ↑ JJ Farmer III, BR Davis u. a .: Biochemical identification of new species and biogroups of Enterobacteriaceae isolated from clinical specimens . In: Journal of Clinical Microbiology . tape 21 , no. 1 , January 1985, p. 46-76 , PMID 3881471 , PMC 271578 (free full text).