Pyrimidine de novo synthesis
| Parent |
|
Nucleoside Synthesis Nucleotide Synthesis |
| Subordinate |
|
UMP synthesis CMP synthesis TMP synthesis |
| Gene Ontology |
|---|
| QuickGO |
The de novo synthesis of pyrimidine is the biochemical synthesis of pyrimidine nucleotides from simpler molecules. In contrast to the salvage pathway , no nucleotides or nucleotide derivatives are converted and reused here, but are rebuilt with greater energy expenditure. In the de novo produced synthesis ribonucleotides , but not deoxyribonucleotides .
The course of the de novo synthesis is highly conserved .
Synthetic route
In the de novo synthesis of pyrimidine, the pyrimidine base is first built up and then placed on a ribose phosphate so that finally a pyrimidine nucleotide is formed. All but one step take place in the cytosol.
Carbamoyl Phosphate Synthesis (Step 1)
Hydrogen and ammonia form the starting materials, the enzyme from carbamoyl phosphate synthetase II (CPS2, EC 6.3.5.5 ) under application of two molecules of ATP to carbamoyl be implemented. The ammonia here comes from L - glutamine .
Attachment of aspartate (step 2)
Carbamoyl phosphate ( 2 ) and the amino acid L - aspartate ( 1 ) are converted to N- carbamoyl aspartate ( 3 ), which catalyzes an aspartate transcarbamoylase (ATCase, EC 2.1.3.2 ).
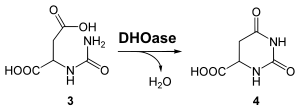
Ring closure (step 3)
The enzyme dihydroorotase ( EC 3.5.2.3 ) catalyzes the intramolecular condensation of carbamoyl aspartate to dihydroorotate ( 4 )
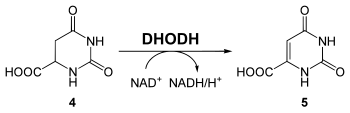
Oxidation to orotate (step 4)
The enzyme dihydroorotate dehydrogenase (DHODH, EC 1.3.3.1 ) oxidizes dihydroorotate to orotate ( 5 ), whereby NAD + as an oxidizing agent is reduced to NADH.
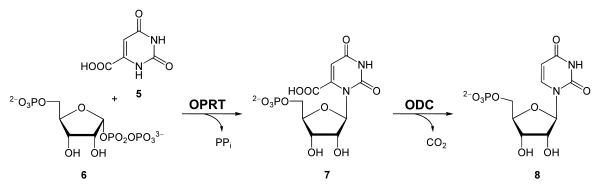
Ribose Transfer and Decarboxylation (Step 5 and 6)
The transfer to a ribose group takes place with the help of orotate phosphoribosyl transferase (OPRTase EC 2.4.2.10 ). Orotate reacts with phosphoribosyl pyrophosphate ( 6 ) with cleavage of pyrophosphate to form orotidine monophosphate (OMP, 7 ). This in turn is of orotidine 5'-phosphate decarboxylase (also Orotidylat decarboxylase, EC 4.1.1.23 ) to uridine monophosphate (UMP, 8 ) decarboxylated .
UMP is the starting product of all other pyrimidine nucleotides such as uridine diphosphate (UDP), UDP-glucose , uridine triphosphate (UTP) and cytidine triphosphate (CTP). Deoxyribonucleotides, building blocks in DNA, are obtained from the corresponding ribonucleotides.
Genomic organization in different species
Although de novo synthesis is practically ubiquitous, there are differences between the species in the organization of the enzymes and the genes coding for them . The number of genes involved in the six enzymatic steps decreases from prokaryotes to eukaryotes , but the complexity of the enzymes increases in the same way. So-called cluster genes are formed , which code for multifunctional enzymes .
Prokaryotes
| gene | enzyme | Surname |
|---|---|---|
| pyrA | CPSase | Carbamoyl phosphate synthetase |
| pyrB | ATCase | Aspartate transcarbamoylase |
| pyrC | DHOase | Dihydroorotase |
| pyrD | DHODH | Dihydroorotate dehydrogenase |
| pyrE | OPRTase | Orotate phosphoribosyl transferase |
| pyrF | ODCase | Orotidine 5'-phosphate decarboxylase |
In bacteria, six different genes ( pyrA-pyrF ) code for six independent enzymes. The bacterium Escherichia coli has a special feature . Its gene pyrA equivalent genes carA and carB coding for the two subunits of the first enzyme CPSase.
Lower eukaryotes
| gene | enzyme | Surname |
|---|---|---|
| ura2 | CPSase | Carbamoyl phosphate synthetase |
| ura2 | ATCase | Aspartate transcarbamoylase |
| ura4 | DHOase | Dihydroorotase |
| ura1 | DHODH | Dihydroorotate dehydrogenase |
| ura5 ura10 | OPRTase | Orotate phosphoribosyl transferase |
| ura3 | ODCase | Orotidine 5'-phosphate decarboxylase |
In Saccharomyces cerevisiae , the ura2 gene codes for an enzyme with bifunctional CPSase-ATCase activity. This enzyme could also be isolated from Neurospora crassa and Aspergillus . The following four enzymatic steps are encoded by the independent genes ura4 (DHOase), ura1 (DHODH), ura5; ura10 (OPRTase) and ura3 (ODCase).
Higher eukaryotes
| gene | enzyme | Surname |
|---|---|---|
| pyr1-3 cad | CPSase | Carbamoyl phosphate synthetase II |
| pyr1-3 cad | ATCase | Aspartate transcarbamoylase |
| pyr1-3 cad | DHOase | Dihydroorotase |
| pyr4 | DHODH | Dihydroorotate dehydrogenase |
| pyr5-6 | OPRTase | Orotate phosphoribosyl transferase |
| pyr5-6 | ODCase | Orotidine 5'-phosphate decarboxylase |
In higher eukaryotes, with the exception of plants, three structural genes are involved in pyrimidine synthesis. A cluster gene pyr1-3 codes for a multifunctional polypeptide, which is responsible for the first three synthesis steps . In mammals it is called cad , derived from CPSase, ATCase, and DHOase. The second gene, pyr4, codes for dihydroorotate dehydrogenase (DHODH). The third is again a cluster gene pyr5-6 and codes for a polypeptide which has OPRTase and ODCase activity.
| gene | enzyme | Surname |
|---|---|---|
| carA / carB | CPSase | Carbamoyl phosphate synthetase |
| pyrB | ATCase | Aspartate transcarbamoylase |
| pyrC | DHOase | Dihydroorotase |
| pyrD | DHODH | Dihydroorotate dehydrogenase |
| pyrE-F | OPRTase | Orotate phosphoribosyl transferase |
| pyrE-F | ODCase | Orotidine 5'-phosphate decarboxylase |
Pyrimidine synthesis in plants also shows a tendency towards the formation of fusion genes, but there is no evidence of a multifunctional complex that catalyzes more than one of the first four enzymatic steps. The genes are called carA / carB (CPSase), pyrB (ATCase), pyrC (DHOase) and pyrD (DHODH). The conversion of orotate to UMP takes place in plants by UMP synthase ( pyrE-F ), a dimeric polypeptide whose monomers each have both enzyme activities of OPRTase and ODCase.
When looking at the fully sequenced genome of Arabidopsis thaliana , it is noticeable that there is only one gene locus for each enzyme in de novo synthesis.
Individual evidence
- ^ KG Wagner, AI Backer: Dynamics of nucleotides in plants studied on a cellular basis. In: Int. Rev. Cytol. 134, 1992, p. 184; doi: 10.1016 / S0074-7696 (08) 62027-6 .
- ↑ M. Denis-Duphil: Pyrimidine biosynthesis in Saccharomyces cerevisiae: the ura2 cluster gene, its multifunctional enzyme product, and other structural or regulatory genes involved in de novo UMP synthesis. In: Biochem. Cell Biol. 67, No. 9, 1989, pp. 612-631.
- ↑ GA O'Donovan, J. Neuhard: pyrimidine metabolism in microorganisms. In: Bacteriol. Rev. 34, No. 3, 1970, 278-343.
- ↑ F. Lacroute: Regulation of pyrimidine biosynthesis in Saccharomyces cerevisiae. In: J. Bacteriol. 95, 1968 pp. 824-832.
- ↑ L. Williams, S Bernhardt; Davis, R .: Copurification of pyrimidine-specific carbamyl phosphate synthetase and aspartate transcarbamylase of Neurospora crassa. In: Biochemistry. 9, No. 22, 1970, pp. 4329-4335.
- ^ L Palmer; Cove, D .: Pyrimidine biosynthesis in Aspergillus nidulans: isolation and preliminary characterization of auxotrophic mutants . In: Mol Gen Genet . 138, No. 3, 1975, pp. 243-255.
- ↑ Jones, 1980.
- ↑ M. Denis-Duphil: Pyrimidine biosynthesis in Saccharomyces cerevisiae: the ura2 cluster gene, its multifunctional enzyme product, and other structural or regulatory genes involved in de novo UMP synthesis. In: Biochem. Cell Biol. 67, No. 9, 1889, pp. 612-631.
- ↑ Walther et al., 1984.
- ^ Arabidopsis Genome Initiative, 2000.
- ↑ R. Boldt, R. Zrenner: Purine and pyrimidine biosynthesis in higher plants . In: Physiol Plant 117, No. 3, 2003, pp. 297-304.